Glucose oxidase-expressing fructooligosaccharide-synthesizing engineered strain, and its construction method and application
A technology of glucose oxidase and fructooligosaccharides, which is applied in the field of genetic engineering, can solve the problems of high production cost of high-purity fructooligosaccharides, unsuitable for large-scale industrial production, complicated operation and control, etc., and achieves strong application value. , reduce the inhibitory effect, and promote the effect of positive transformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
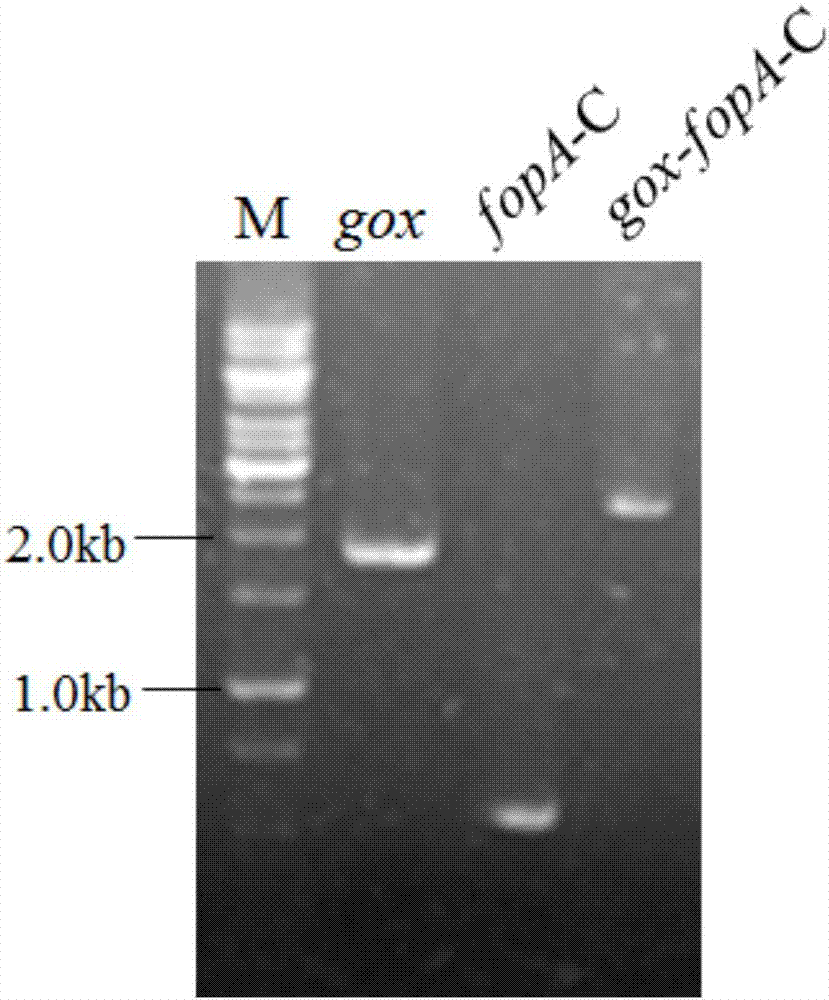
[0030] Example 1. The fusion of the C-terminal domain gene fopA-C of the glucose oxidase gene gox and the β-fructofuranosidase FopA to obtain the fusion gene gox-fopA-C
[0031]First, use Aspergillus niger ATCC1015 chromosomal DNA as a template, gox-1F / gox-1815R as primers to amplify a 1.8kb gox gene fragment with a fopA-C linker without a stop codon, and use Aspergillus niger ATCC 20611 chromosomal DNA as a template , fopA-438F / fopA-965R are primers to amplify the fopA-C gene fragment of 0.3kb, and then use Double jointPCR technology to fuse gox and fopA-C two genes to obtain a gox-fopA-C fusion gene with a size of 2.1kb ( figure 1 ).
[0032] The above primer pairs are as follows:
[0033] gox-1F: ATGCAGACTCTCCTTGTGAG
[0034] gox-1815R: CTGAACTGGTAGTAGATGGCCTGCATGGAAGCATAATC
[0035] fopA-438F: GCCATCTACTACCAGTTC
[0036] fopA-965R: TCACCACGATCTCGCCCAGGT
Embodiment 2
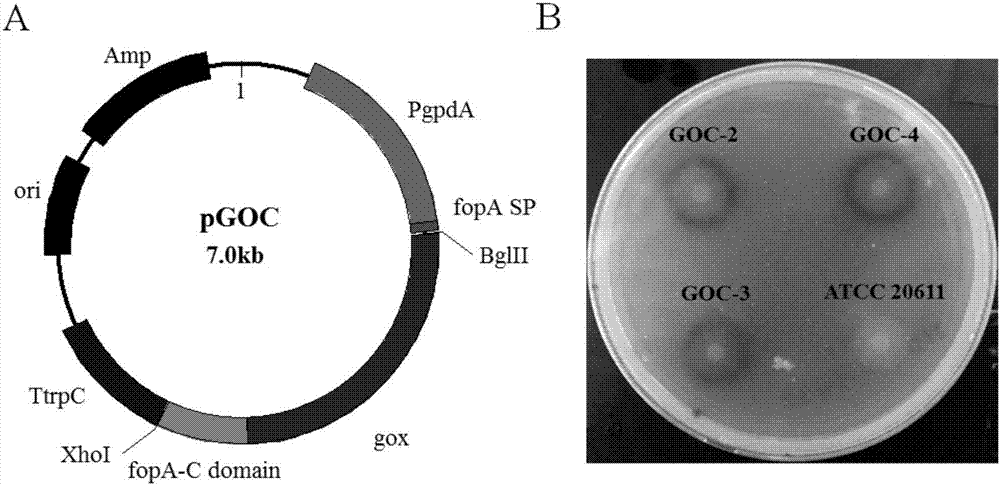
[0037] Example 2. Construction of glucose oxidase GOX fusion FopA C-terminal domain expression vector pGOC
[0038] The expression vector pGOC was constructed with the pMD 19-T vector as the backbone, including the C-terminal domain fusion gene gox-fopA-C of the promoter PgpdA, glucose oxidase and FopA, and the terminator TtrpC ( figure 2 A); First, using the plasmid pAN7-1 (Punt et al., Gene.1987, 56, 117-24.) as a template, the primer pair PgpdA-1192UF / PgpdA-7UR and TtrpC-1044DF / TtrpC-1805DR were used to amplify gox The 1.2kb PgpdA of the joint and the TtrpC with the fopA-C joint of 0.7kb, then the fusion gene gox-fopA-C is connected with the promoter PgpdA and the terminator TtrpC to form the fusion gene gox-fopA-C expression cassette; The fusion fragment of the expression cassette was cloned into the pMD 19-T vector, and the single clone was picked for verification. The verified plasmid was named pGOC. See the plasmid map figure 2 As shown in A.
[0039] The above prim...
Embodiment 3
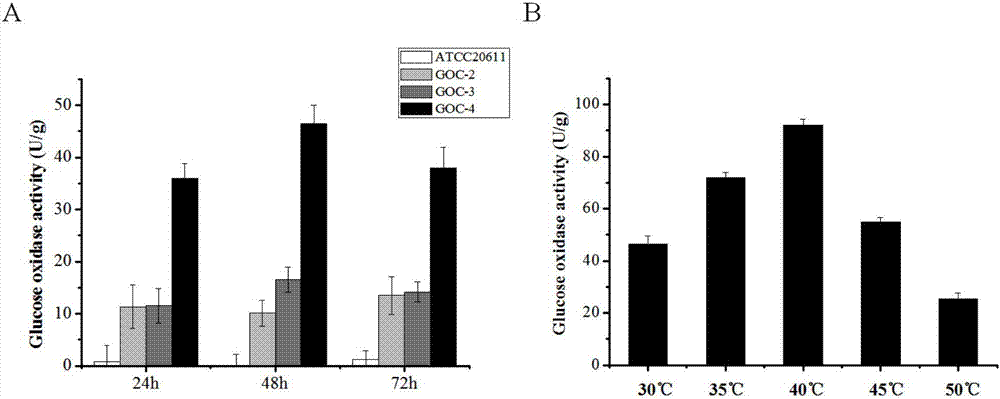
[0044] Example 3. Using expression vector pGOC to transform Aspergillus niger ATCC 20611 to construct fructooligosaccharide synthesis engineering bacteria expressing glucose oxidase
[0045] The genetic transformation of Aspergillus niger ATCC 20611 was performed using PEG / CaCl 2 Mediated protoplast transformation method, pyrithione resistance gene ptrA as selection marker. 10 μg of the purified plasmid pGOC and plasmid pME2892 (Krappmann et al., Eukaryotic Cell. 2005, 4, 1298–1307.) containing the ptrA expression cassette were mixed and co-transformed into Aspergillus niger ATCC 20611.
[0046] The above PEG / CaCl 2 The specific method of mediated protoplast transformation is as follows:
[0047] (1) Preparation of Aspergillus niger ATCC 20611 protoplasts:
[0048] Aspergillus niger ATCC 20611 spores were inoculated into 50ml MM liquid medium (the final concentration of spores was 1×10 6 Individual / ml), 30 ℃, 200rpm culture 30h; The thalline is collected by centrifugation ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com