Nucleic acid mass spectrometry method for detecting 10 common pathogenic bacteria of clinical infection
A technology for detection of pathogenic bacteria and pathogenic bacteria, applied in the direction of microorganism-based methods, biochemical equipment and methods, and measurement/inspection of microorganisms, can solve problems such as inability to meet the clinical needs of various pathogenic microorganisms, and achieve a high degree of automation and high detection sensitivity High, easy and safe operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] Example 1: Primer Design and Synthesis
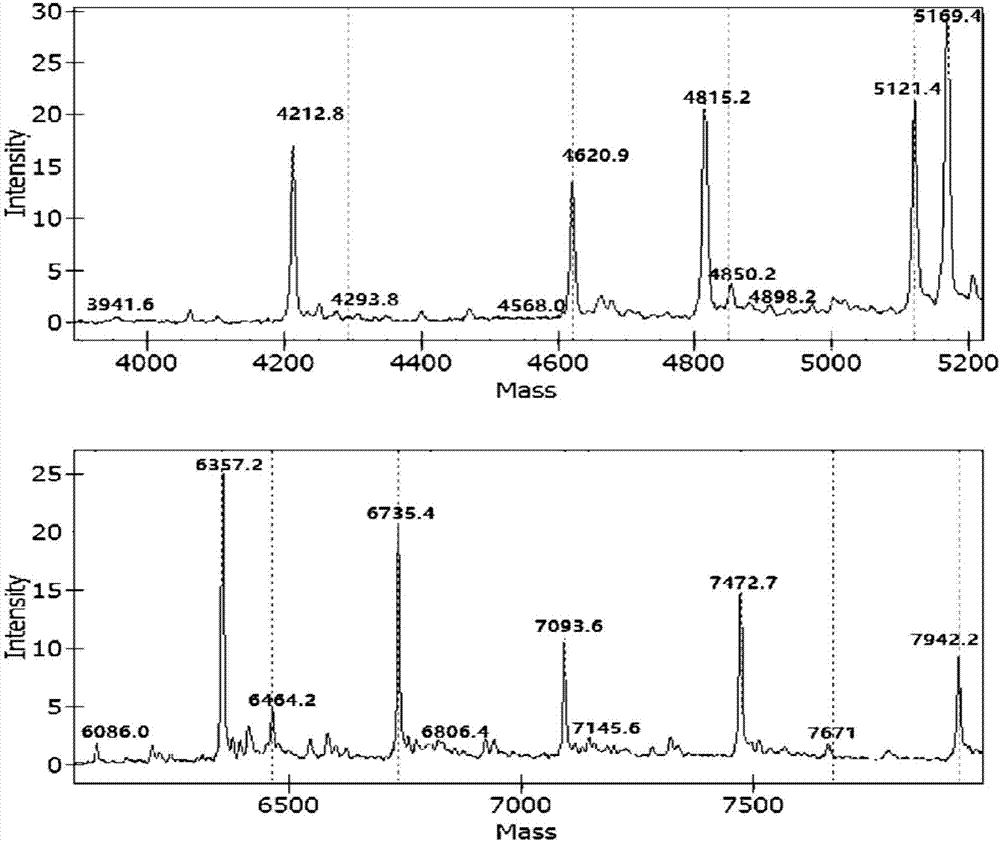
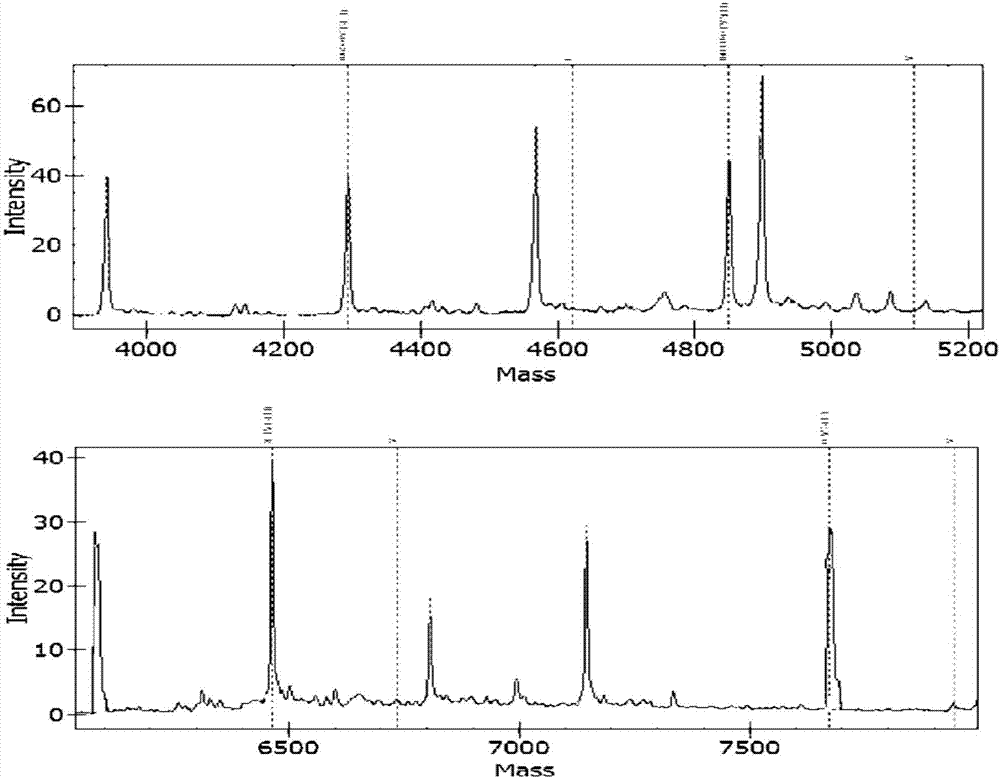
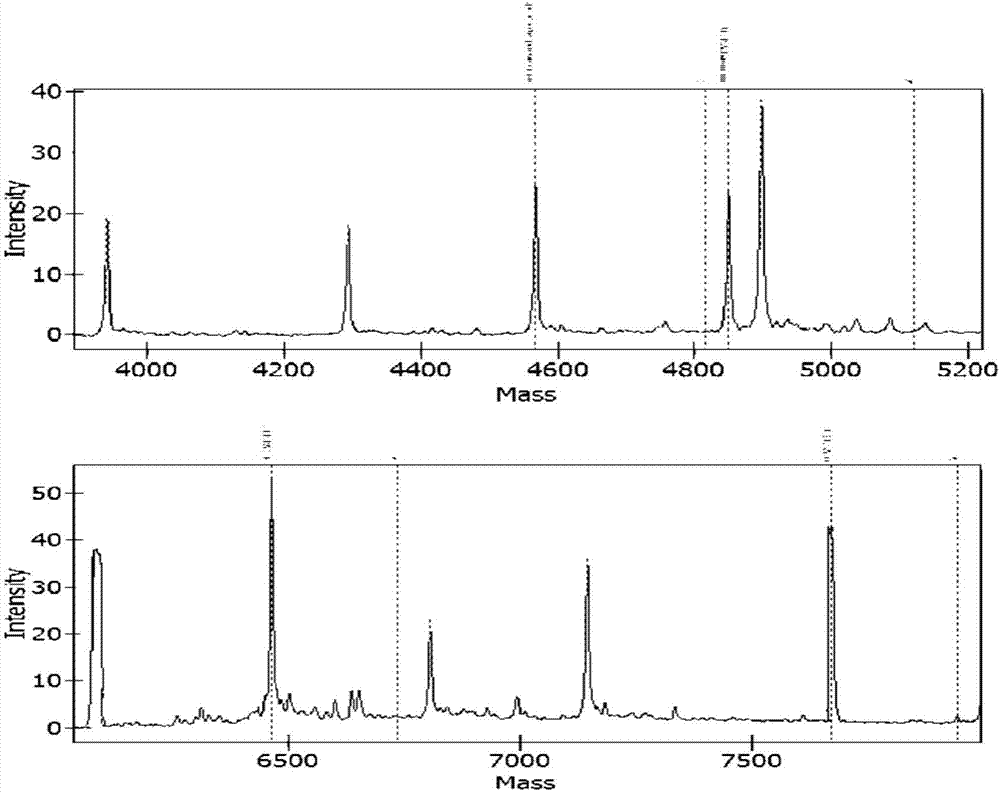
[0095] Targeting 10 virulence factor genes related to pathogenic bacteria: Klebsiella pneumonia virulence factor khe gene, Acinetobacter baumannii virulence factor OXA gene, Enterococcus faecium virulence factor ddl gene, Streptococcus pneumonia virulence factor Lyt gene, Enterococcus faecalis virulence factor ddl gene, Escherichia coli virulence factor phoA gene, Staphylococcus aureus virulence factor nuc gene, Proteus mirabilis virulence factor ureR gene, Stagphylococcus epidermidis virulence factor nuc gene, Pseudomonas aeruginosa virulence factor ecfX gene related specific sites, design corresponding Specific PCR primer core sequence (SEQ ID No: 1 to SEQ ID No: 20) and specific extension primer core sequence (SEQ ID No: 21 to SEQ ID No: 30).
[0096] Among them, in order to prevent the PCR primers from entering the detection window of the mass spectrometer and interfering with the detection effect, a certain number of bases c...
Embodiment 2
[0099] Embodiment 2: sample DNA extraction
[0100] Collect samples from patients with clinical pathogen infection (hot (hot stained samples or low temperature hot stained samples with chills, leukocytosis (count greater than 10.0) 9 / L, especially when there is "nuclear shift to the left", skin and mucous membrane hemorrhage, coma, multiple organ failure, lower blood pressure, higher C-reactive protein and faster breathing, neutropenia and thrombocytopenia in patients with blood diseases, etc., or at the same time With the above signs) blood samples from 6 cases. Among them, DNA extraction and so on are in accordance with the requirements of the instructions, and human venous blood is collected when the patient has chills, fever, and body temperature reaches the highest point, and is collected with EDTA anticoagulant tubes. According to the instructions, the collected blood should not be stored at 2-8°C for more than one week, and at -20°C for no more than one month. Try to ...
Embodiment 3
[0101] Embodiment three: Biological experiment
[0102] The gene loci related to the virulence factor genes of 10 common pathogenic bacteria in clinical infection were tested using the PCR instrument of Yixin Bochuang Company.
[0103] The components used in the detection system for PCR, PCR product purification and single base extension are:
[0104] serial number
component name
main ingredient
1
PCR mix
dNTPs, MgCl2
2
PCR Primer Mix
PCR primers
3
PCR enzyme
Taq enzyme
4
SAP Enzyme Mix
SAP enzyme
5
Extension Primer Mix
extension primer
6
elongase mix
iPLEX enzymes, ddNTPs
7
Genomic DNA of 10 kinds of bacteria (10ng / ul)
[0105] According to the manual, the specific operation method is as follows:
[0106] 1. PCR amplification
[0107] 1.1 In the PCR preparation area, prepare 200ul PCR reaction tubes according to the number of samples to be...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com