Application of miR-21 and target gene thereof in preparation of reagents or drugs diagnosing and/or controlling osteoarthritis
Osteoarthritis, mir-21 technology, applied in anti-inflammatory agents, disease diagnosis, drug combination, etc., can solve the imbalance between chondrocyte synthesis and catabolism, chondrocyte apoptosis, and silencing of PI3K/Akt/mTOR signaling pathway Questions such as ambiguity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
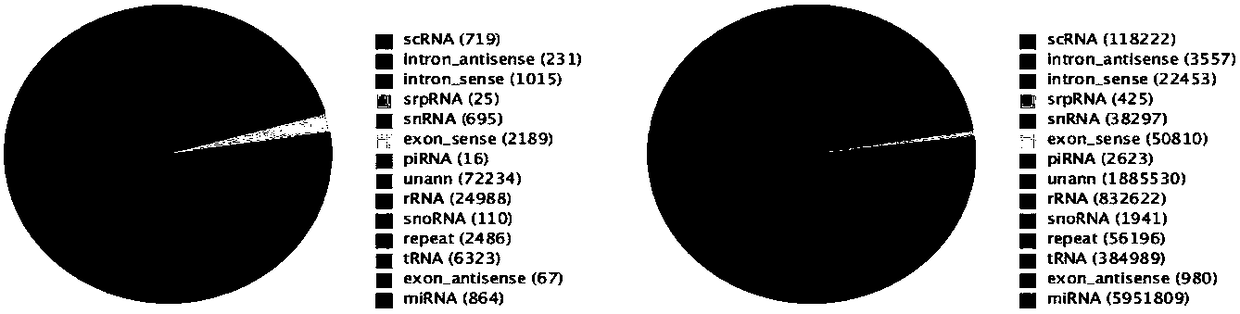
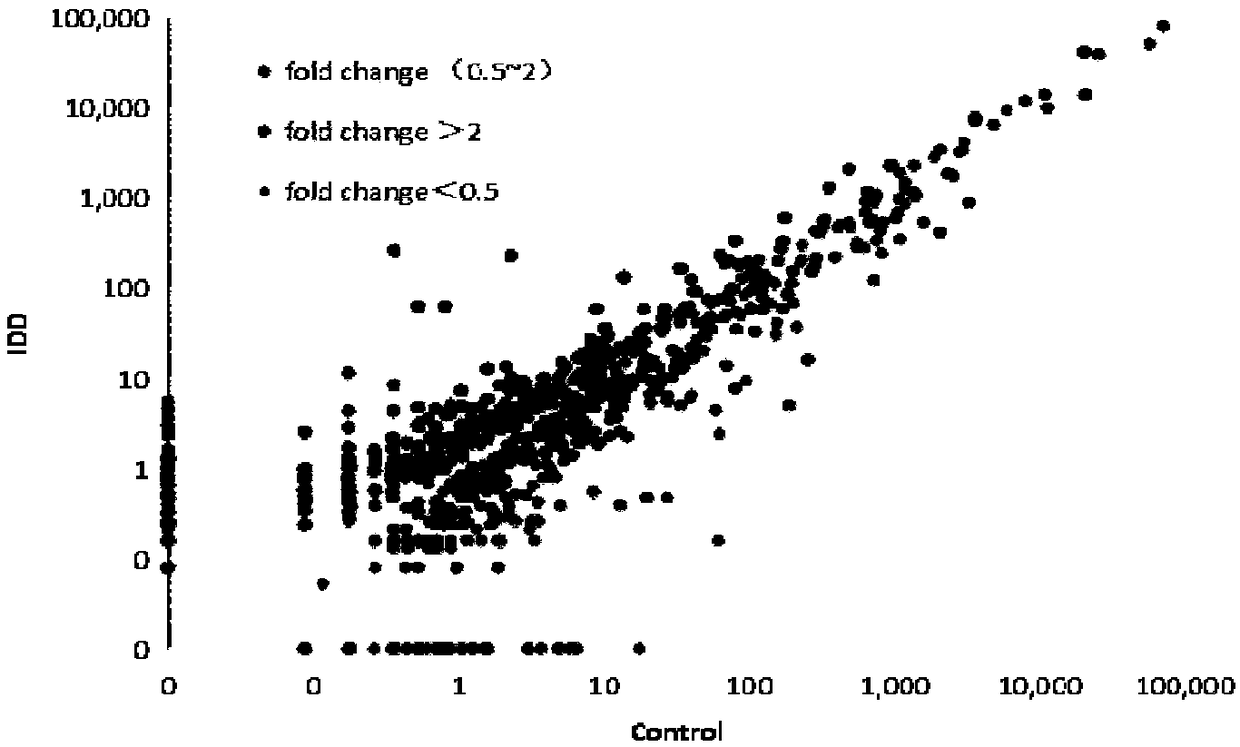
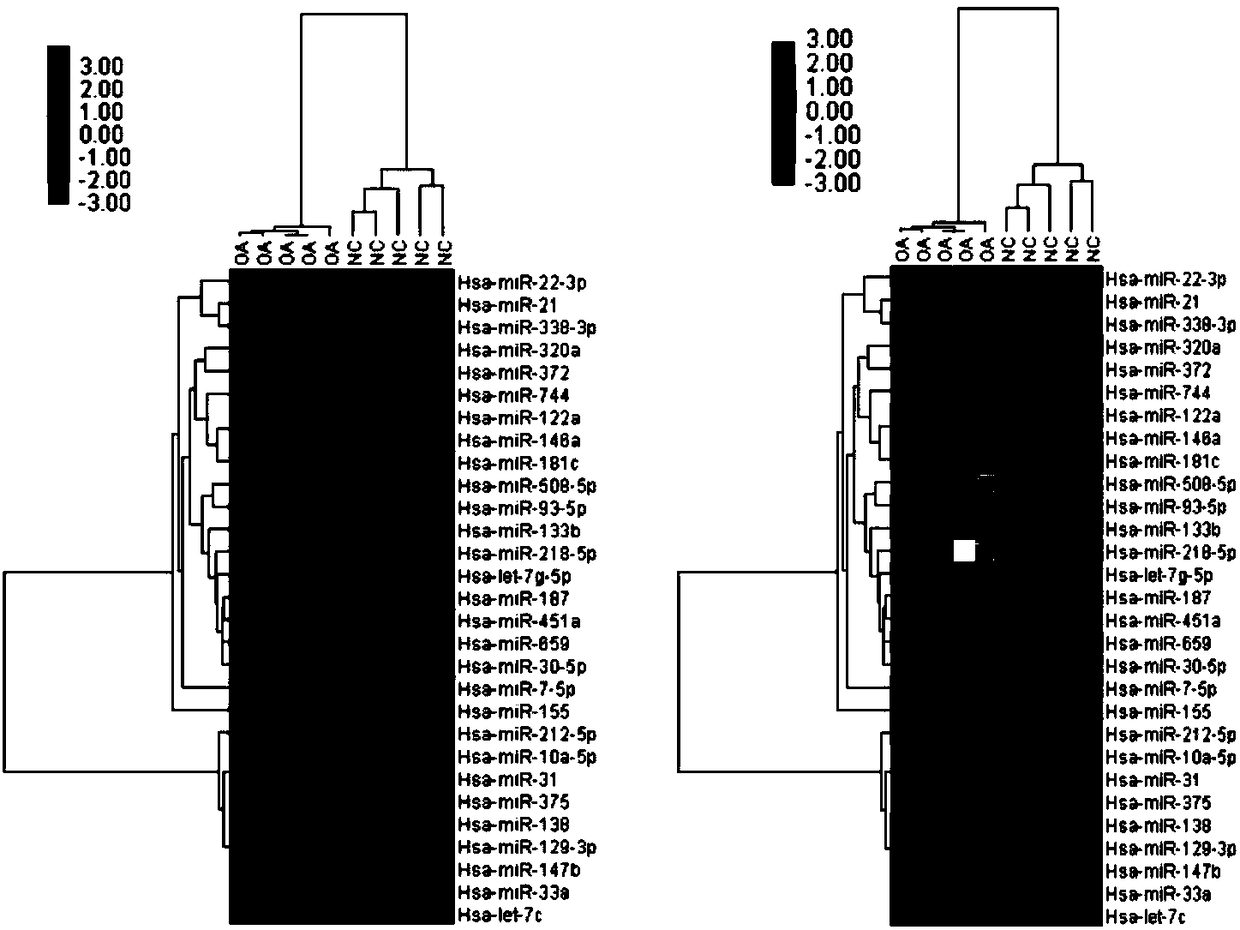
[0048] Example 1: Detection of miRNA expression profile in cartilage tissue of patients with osteoarthritis and normal control group
[0049] Experimental objects: case group (knee cartilage of patients with osteoarthritis) and control group (knee cartilage of patients with amputation).
[0050] 1. Total RNA extraction
[0051] After informed consent was obtained from the subjects, the joint fluid was extracted through articular cavity puncture, placed in sterile, enzyme-free cryopreservation tubes, and stored in liquid nitrogen.
[0052] The total RNA was extracted by the Trizol one-step method, and the specific operation was as follows: take 300 μL of joint fluid, add 4 times the volume of Trizol solution (TRIzol Reagent, Invitrogen, USA, Cat. No. 15596-018), then add 200 μL of chloroform, shake the centrifuge tube vigorously, and mix well. Place at room temperature for 5-10min; centrifuge at 12000rpm for 15min and transfer the upper aqueous phase to another new centrifuge ...
Embodiment 2
[0072] Example 2: Bioinformatics method predicts that PIK3C2A gene is identified as the target gene of miR-21
[0073] Apply bioinformatics methods to predict the target gene, by comparing the matching degree of miRNA and the 3'-UTR of the target gene, especially the 6-8 bp seed sequence at the 5' end of miRNA. The prediction of miRNA target molecules is mainly based on the principle of base pairing, and the target gene is predicted according to the complementarity between the 3'-UTR of the target molecule and the miRNA through the bioinformatics database. False-positive and false-negative results are unavoidable if any target molecule database is used alone, so four independent microRNA target molecule databases are used, namely miRanda (http: / / www.microrna.org / ), TargetScan (http: / / www.targetscan.org / ), PicTar (http: / / pictar.mdc-berlin.de / ) and PITA (http: / / genie.weizmann.ac.il / pubs / mir07 / mir07), predicted miR-21 The target gene is PIK3C2A.
Embodiment 3
[0074] Example 3: Construction of PIK3C2A wild-type and mutant expression vectors
[0075] In order to further verify the theoretical target gene, the expression vectors of PIK3C2A wild type (wild type) and mutation prediction sites (mutant type, MT) were constructed, and then the expression vectors were inserted into the luciferase reporter gene vector (pmiR- REPORT, Ambion, USA), psi-CHECK-PIK3C2A WT and psi-CHECK-PIK3C2A MT were determined by DNA sequencing. Lipofectamine 3000 was used to transfect PIK3C2A3'-UTR WT and MT recombinant vectors into chondrocytes respectively, each vector blank group, transfected miR-21mimic group, mimic negative control group, miR-21inhibitor group, inhibitor negative control group, and continued to culture cells for 24 hours According to the instructions of Promega's dual luciferase detection kit, three replicate wells were set up in each group, and the fluorescence intensity of cells in each well of the 24-well plate was detected. The ratio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com