Construction, expression and application of tumor-targeted peptide F3 modified cowpea chlorotic mottle virus-like particles
A chlorotic mottle virus and tumor targeting technology, applied in the field of genetic engineering, can solve the problems of lipophilicity and limited application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Construction of recombinant expression plasmid pPICZ A-F3-CP (ΔN26)
[0046] The tumor-targeting peptide F3 gene was inserted into the CCMVCP (ΔN26) gene with 1-26 amino acid residues deleted at the amino terminal by PCR method to construct the fusion gene F3-CP (ΔN26). The 1-25 amino acid residues at the amino terminus of CCMV CP are the RNA binding domain, and studies have shown that its knockout or replacement does not affect the assembly of CP dimers into VLPs. First, two sets of DNA oligonucleotides Olig-1 and Olig-C1, Olig-2 and Olig-C2 with complementary sequences were designed and annealed to obtain F3 gene and CP(ΔN26) fragments respectively, with 50 overlapping bases between them Base (Table 1). Subsequently, double asymmetric PCR (DA-PCR) and overlap extension PCR (OE-PCR) were used for two-step gene synthesis with corresponding primers (F3-Up and F3-Dn, CP-Up and CP-Dn). The overlapping bases between them serve as bridges. Finally, the fusion ge...
Embodiment 2F3
[0053] Example 2 Induced expression and purification of F3-CCMV virus-like particles
[0054] After linearizing the recombinant plasmid pPICZ A-F3-CP (ΔN26) with SacI, it was transformed into Pichia pastoris GS115 by electroporation. The transformed Pichia pastoris was spread on a YPD plate (1% yeast extract, 2% peptone, 2% glucose, 2% agar powder) containing 100 μg / mL bleomycin Zeocin, cultured upside down at 30°C for 2-3 days, and screened positive transformants.
[0055] The identified positive transformants were inoculated with buffered glycerol complex medium BMGY and buffered methanol complex medium for methanol induction. Collect yeast cells by centrifugation, add an equal volume of 0.5mm acid-washed glass beads (purchased from Sigma) and 5 times the volume of breaking buffer (pH 5.5, 50mM sodium phosphate, 1mM PMSF, 1mM EDTA, 5% glycerol), and vortex broken. Centrifuge to collect the supernatant, carry out protein gel electrophoresis (SDS-PAGE), detect the expressio...
Embodiment 3F3
[0057] Identification of embodiment 3F3-CCMV virus-like particles
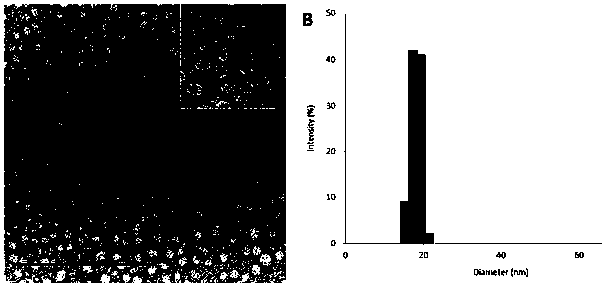
[0058] The morphology of the purified F3-CCMV virus-like particles was observed under a transmission electron microscope (nickel mesh, negatively stained with 2% uranyl acetate). virus particles (such as image 3 shown in A). Dynamic light scattering DLS determines that its hydrodynamic diameter is 18.2 ± 1.4nm, has good monodispersity (see image 3 B). This and the CCMV capsid protein CP in the prior art are tagged with histidine (His 6 ) and elastin-like polypeptide (ELP) modified virus-like particles are similar in size, and should be pseudovirus particles composed of 120 CP subunits (T=2), rather than wild CCMV composed of 180 CP subunits Virus particles (T=3).
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com