alpha-L-arabinofuranosidase capable of improving filtration performance of wort
A furanosidase and furanosidase technology, which is applied in the field of bioengineering, can solve the problems of few AnabfA application research and insufficient AnabfA attention, and achieves the effect of improving the filtration speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Analysis and cloning of Aspergillus niger α-L-arabinofuranosidase gene
[0038] The genome of Aspergillus niger was extracted by grinding with liquid nitrogen. The gene size of the Aspergillus niger arabinofuranosidase query from Genbank is 1790bp, and there is only one 50bp intron sequence. The encoded protein sequence contains 579 amino acids. The amino acid sequence of α-L-arabinofuranosidase is predicted by the signal peptide prediction software SignaIP. The results show that the N-terminal of α-L-arabinofuranosidase contains 18 amino acid signal peptides , the theoretical molecular weight of the protein is 70kDa. The glycosylation level and glycosylation sites of the enzyme were analyzed using NetNGlyc 1.0 Server and NetOGlyc 4.0 Server. The results showed that there were 9 N glycosylation sites on the amino acid strip of recombinant α-L-arabinofuranosidase and 5 O-glycosylation sites.
[0039] Primers P1, P2, P3, and P4 were designed, and the primer ...
Embodiment 2
[0042] Embodiment 2: Preparation of recombinant α-L-arabinofuranosidase sidase
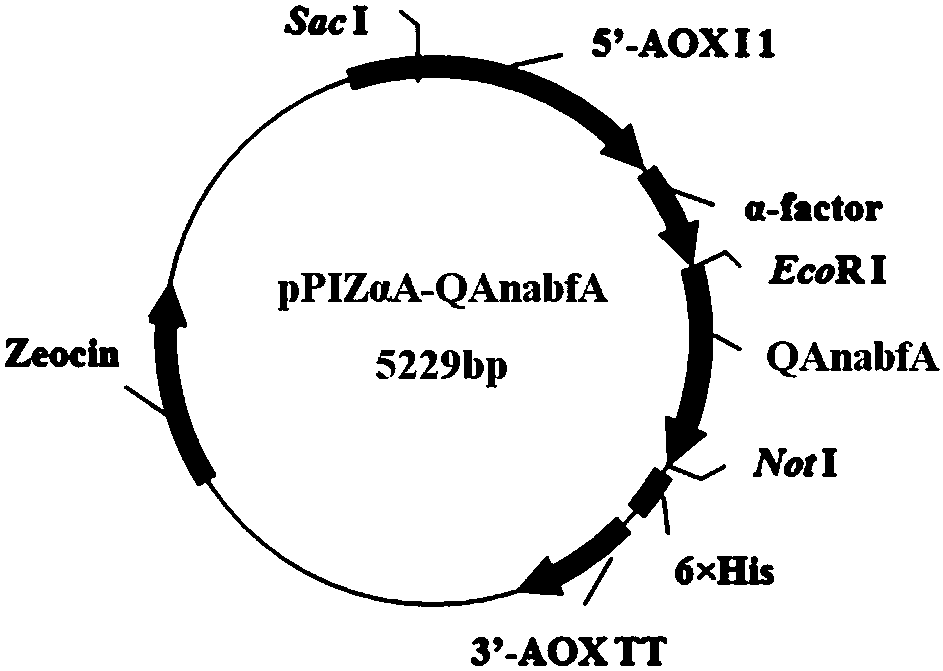
[0043] The α-L-arabinofuranosidase gene fragment (shown in SEQID NO.2) with the signal peptide, intron and stop codon sequence removed was connected to the expression vector to obtain the recombinant expression plasmid pPICZαA-QAnabfA, and the recombinant expression plasmid pPIZαA -The construction map of QAnabfA is as figure 1 shown. After the recombinant expression vector pPICZαA-QAnabfA was linearized by Sac I digestion, it was transformed into Pichia pastoris X-33 by electroporation, and the transformed Pichia pastoris was initially screened with a plate containing zeocin antibiotic to obtain positive transformants . The genome was extracted, and the positive transformants were screened again by PCR to obtain recombinant strains.
[0044] BMGY was used to expand the seeds of recombinant strains and no-load recombinant strains, and BMMY medium was used to ferment and cultivate the recombinan...
Embodiment 3
[0047] Example 3 Enzyme Activity Determination and Enzymatic Property Analysis of Recombinant α-L-arabinofuranosidase
[0048] The reaction system for the determination of enzyme activity is 200 μl, take 25 μl of p-nitrophenyl arabinofuranoside solution, 50 μl of 0.1M pH5.5 sodium acetate buffer, and 25 μl of enzyme solution in a water bath at 50°C for 15 minutes, and inactivate it with a boiling water bath for 10 minutes. The enzyme solution was used as a blank control, and 100 μl of 1M Na was added quickly after the reaction 2 CO 3 Terminate the reaction, after standing at room temperature for 15 minutes, measure the OD value at 410 nm with a microplate reader, and calculate the content of p-nitrophenol in the sample according to the standard curve prepared by p-nitrophenol. Definition of enzyme activity: the amount of enzyme required to produce 1 μM p-nitrophenol per minute is one enzyme unit. The concentration of the protein in the fermentation broth was determined by Br...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com