Nucleic acid aptamer capable of being specifically combined with carapace arginine kinase, kit and detection method
A technology of arginine kinase and nucleic acid aptamer, which is applied in the field of food analysis, can solve the problems of strict storage conditions, high price of monoclonal antibody, and cumbersome preparation, and achieve high sensitivity, avoid false positives, and simple chemical modification.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Preparation of Arginine Kinase: (1) Take 50 g of Penaeus vannamei muscle, remove the head, tail, shell and gut. (2) Shrimp muscle was cut into mud with a knife, dissolved in buffer A (50mM NaCl, 2mM NaHCO 3 , 10mM EDTA) were homogenized with a homogenizer and allowed to stand at 4°C for 2h. (3) 8000r / min, centrifuge at 4°C for 20min, take the supernatant, add 70% ammonium sulfate, and let stand at 4°C for 8h. (4) 8000r / min, centrifuge at 4°C for 20min, take the supernatant, add 90% ammonium sulfate, and let stand at 4°C for 6h. (5) 8000r / min, centrifuge at 4°C for 20min, and take the precipitate. The precipitate was dissolved in Buffer B (20 mM Tris-HCL, 1 mM NaCl, pH 8.0). (6) ANX Sepharose Fast Flow anion exchange column was used for gradient elution with 0.5M NaCl solution, and the eluted product was collected to obtain arginine kinase for future use.
Embodiment 2
[0033] Screening of nucleic acid aptamers.
[0034] Construction of random ssDNA library of crustacean arginine kinase. (Library capacity is 10 14 The fragment length is 40bp, commissioned by Sangon Bioengineering (Shanghai) Co., Ltd.)
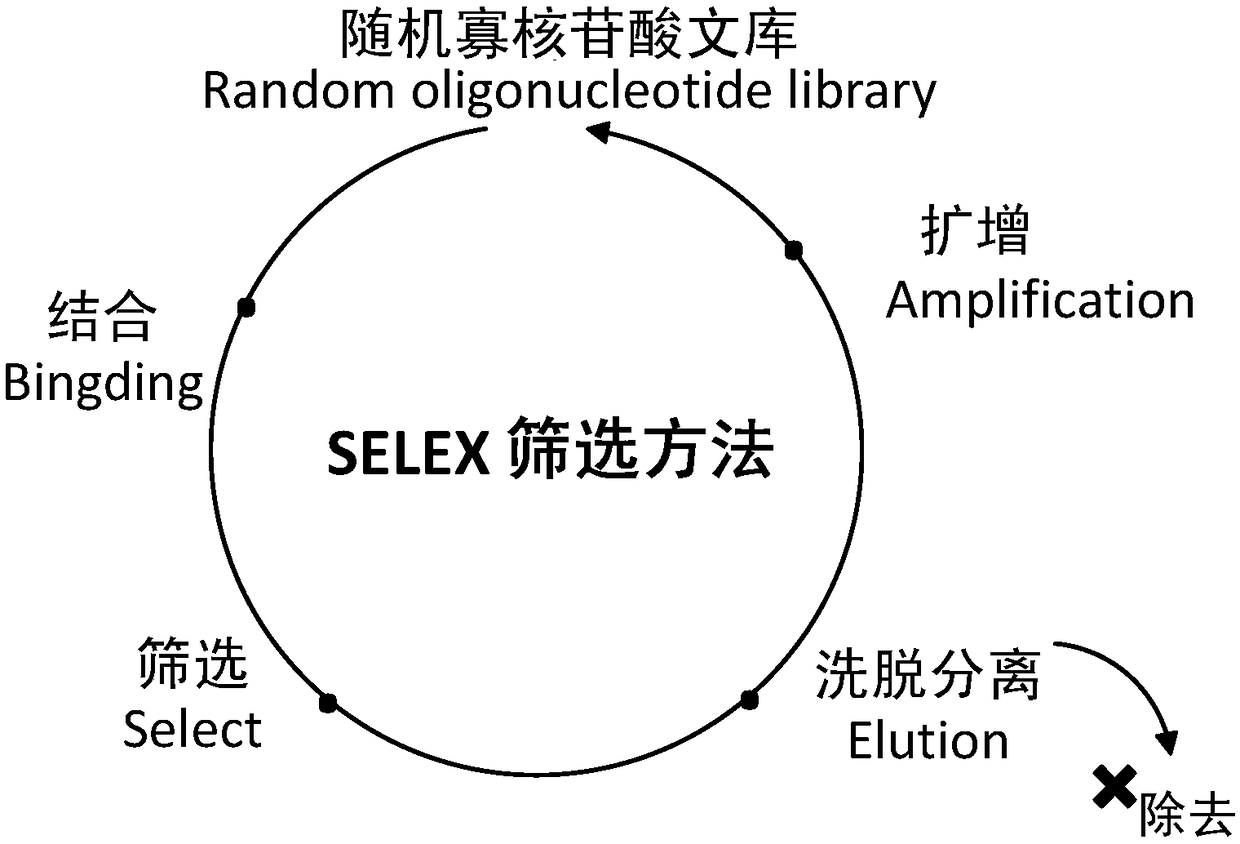
[0035] Such as figure 1 As shown, the present invention adopts three modes of alternate screening.
[0036] In the first screening, the ssDNA library was incubated with arginine kinase (prepared in Example 1) at 37° C. for 1 h, and then added to graphene solution (graphene powder, 5 mg / ml, purchased from Aladdin Reagent (Shanghai) Co., Ltd. company). The resulting mixture was incubated at 37°C to adsorb free-loose ssDNA. The mixture was centrifuged at 12,000 rpm for 15 min to separate and remove unbound ssDNA and graphene in the precipitate, and the remaining supernatant contained ssDNA bound to arginine kinase for asymmetric PCR amplification (restriction primer The ratio of non-restrictive primers is 1:50; the sequences are 5'-CAG GGG ...
Embodiment 3
[0042] First use piraha solution (H 2 SO 4 :H 2 o 2 =3:1) Soak the optical fiber probe with 600 μm quartz fiber (NA=0.22, Nanjing Chunhui) for 30 min, wash it thoroughly with ultrapure water, and dry it with nitrogen to make the surface of the probe hydroxylated; then soak it in 2% (v / v) toluene solution Clean the fiber optic probe for 1 hour with toluene and dry it with nitrogen, then soak the fiber optic probe with 5% (v / v) glutaraldehyde solution at 37°C for 2 hours, wash it with toluene, and dry it in an oven at 120°C to silanize the surface of the probe ; Soak the treated fiber optic probe in arginine kinase solution (concentration: 1mg / mL), soak overnight at 4°C to connect the coated arginine kinase; rinse the fiber optic probe with ultrapure water, and then place it in a 4mg / mL BSA (bovine serum albumin) solution for 2 h to block non-specific adsorption sites. Coated fiber optic probes can be stored at 4°C for several months.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com