Microbial agent containing halophilic denitrifying bacteria YL5-2 and application of microbial agent
A technology of microbial agent and denitrifying bacteria, which is applied in the field of microbial agent containing halophilic denitrifying bacteria YL5-2, can solve the problem of less halophilic denitrifying bacteria, achieve nitrogen nutrient consumption and inhibit algal overgrowth , cost reduction effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Separation and preservation of the halophilic denitrifying bacteria
[0032] The halophilic denitrifying bacteria YL5-2 was isolated from the sedimentary soil of Qarhan Salt Lake (36°51′N, 94°95′E) in Golmud, Qinghai Province, China. The water of the Chaerhan Salt Lake is saturated or close to the saturated salt concentration all the year round.
[0033]LB liquid medium with NaCl concentration of 20% was prepared, and 250 mg / L of glycerol, 250 mg / L of glucose and 50 mg / L of methanol were added, and the sedimentary soil of Chaerhan Salt Lake was enriched and cultured at 30℃ for 72 hours. The strains in the enriched culture medium were isolated using YL solid medium. 1L medium contains the following components: glucose: 0.6g, trisodium citrate 0.5g, glycerol 2mL, yeast extract 0.8g, peptone 1.6g, dipotassium hydrogen phosphate 0.35g, potassium dihydrogen phosphate 0.1g, ammonium sulfate 0.25g, Ammonium Chloride 0.25g, MgSO 4 0.5g, CaCl 2 0.1g, NaCl 180g; t...
Embodiment 2
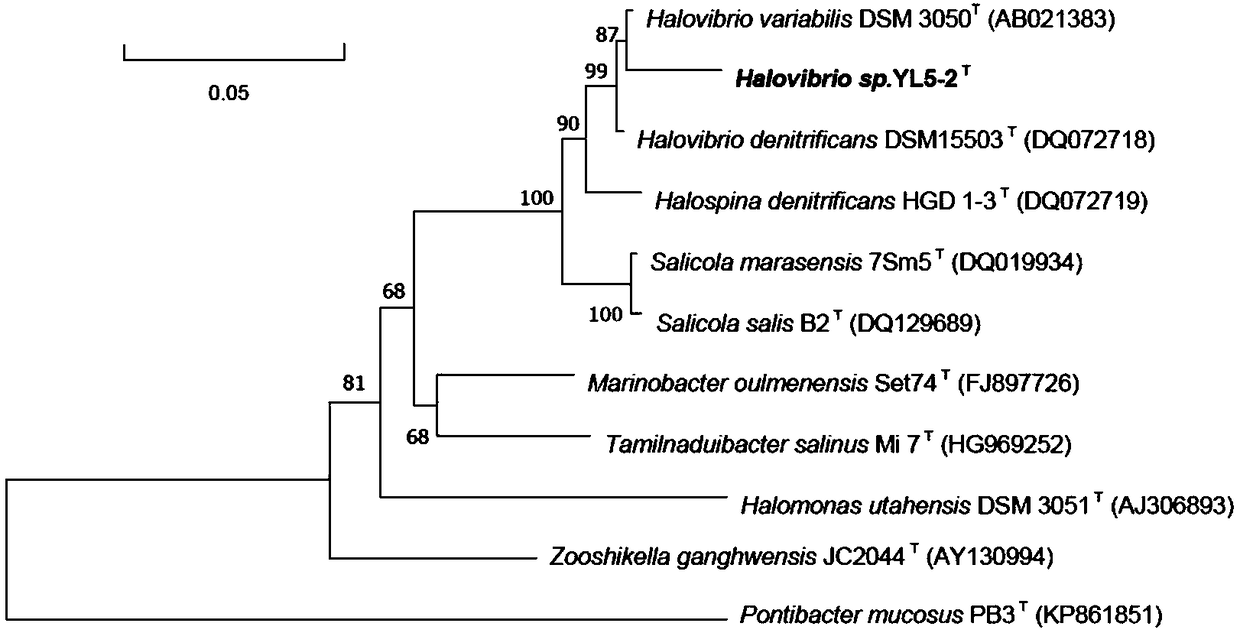
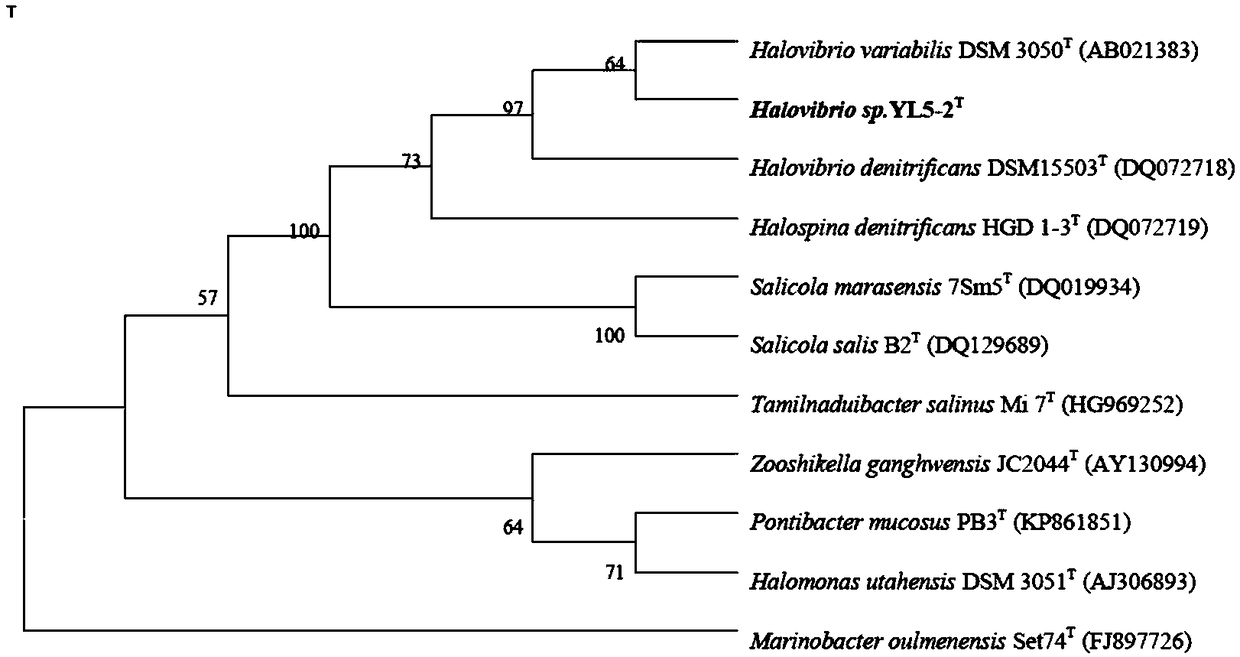
[0035] Example 2 16S rRNA sequence analysis and complete gene sequence analysis of halophilic denitrifying bacteria YL5-2
[0036] The genomic DNA of strain YL5-2 was extracted using TaKaRa kit (TaKaRa MiniBEST Bacteria Genomic DNA Extraction 68Kit Ver.3.0).
[0037] 16S rRNA was amplified using universal bacterial primers 27F (5'-AGAGTTTGATCMTGGCTCA G-3') and 1492R (5'-TACGGYTACCTTGTTACGACTT-3'). PCR sequencing was entrusted to Shanghai Sangon Biotechnology Co., Ltd. The complete 16S rRNA sequence of strain YL5-2 is 1518 bp, as shown in SEQ ID NO. 1, and the GenBank accession number is MF782425.
[0038] Whole-genome sequencing of strain YL5-2 was performed using the IlluminaMiSeq 2000 high-throughput sequencing platform of Shanghai Paisenuo Biotechnology Co., Ltd. Raw sequencing data were filtered and corrected using PRINSEQ (version number v 0.20.4) software, followed by base pairing of genomes using SOAP denovo software (version number v1.05) software with default parame...
Embodiment 3
[0041] Example 3 Identification of phenotypic characteristics and physiological and biochemical characteristics of halophilic denitrifying bacteria YL5-2
[0042] Gram staining properties were tested using the BD Gram staining kit.
[0043] Cell motility was determined using half MA medium (0.5% agar, w / v).
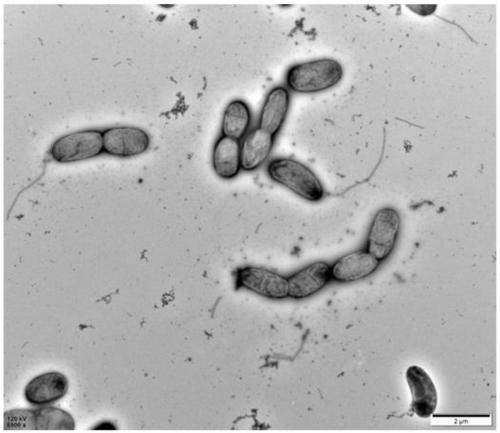
[0044] Cell morphology was detected by transmission electron microscopy (TEM) analysis. That is, cells were picked from the exponentially growing medium, stained with 0.5% uranyl acetate, and photographed under a microscope (Tecnai Spirit, FEI, Hillsboro, OR, USA).
[0045] Oxidase activity was measured using an oxidase kit (bioMérieux) by adding 3.0% H 2 O 2 The solution was poured into bacterial colonies and observed for bubble generation to determine catalase activity.
[0046] The temperature growth conditions were carried out on YL liquid agar medium, the temperatures were 4, 10, 15, 20, 25, 30, 33, 37, 40, 45 and 50 °C, and the pH was constant at 7.5. The strain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com