Oligonucleotide aptamer for specifically recognizing histamine and application thereof
A technology of oligonucleotides and aptamers, which is applied in the field of food safety biology, can solve the problems of time-consuming and laborious, poor stability of recognition molecules, and difficult preparations, and achieve the effects of sensitive detection, short screening cycle, and convenient synthesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: Screening of aptamers
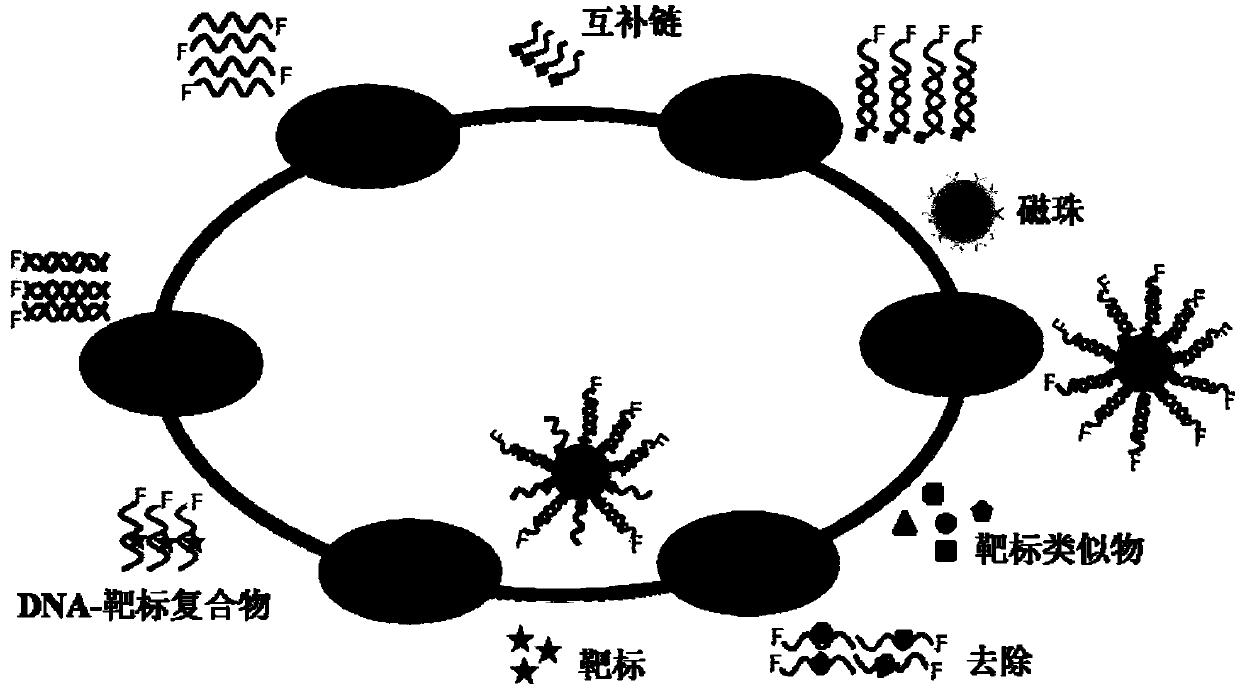
[0022] figure 1 The flow chart of using Capture-SELEX to screen aptamers that can specifically recognize histamine specifically includes the following steps:
[0023] (1) Library hybridization: design a complementary chain CO that is complementary to some bases in the primer region of the library and perform biotin-labeling, and hybridize the ssDNA library to the complementary chain CO;
[0024] (2) Library immobilization: the ssDNA library was immobilized on the surface of the magnetic beads by specific binding of biotin labeled on the complementary chain CO and streptavidin on the magnetic beads, and the non-specifically bound ssDNA was removed by repeated washing;
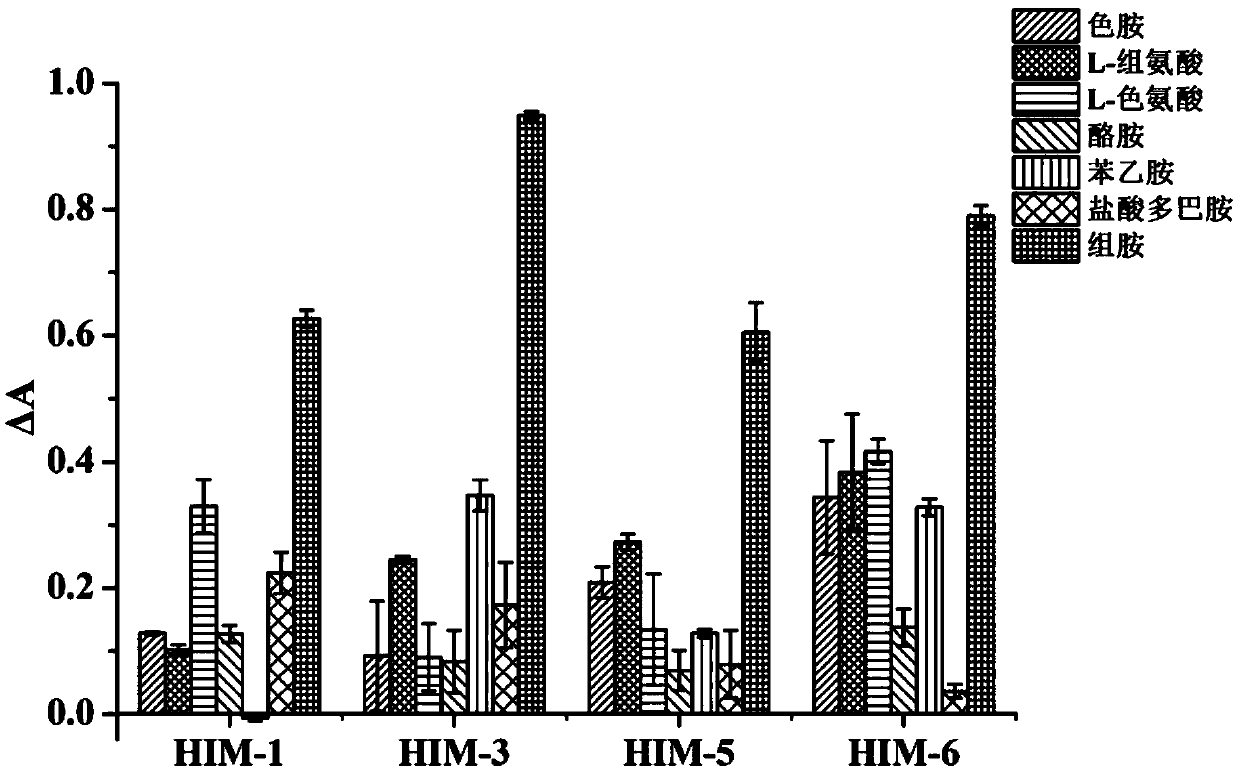
[0025] (3) Anti-screening incubation: During the screening process, in the 4th, 7th, 9th, 12th, 14th, 15th, 16th, and 17th rounds, the structural analogues of histamine, tryptamine, tyramine, phenethylamine, dopamine hydrochloride, L-Histidine and L-Tryptophan are pre-...
Embodiment 2
[0032] Example 2: Cloning and sequencing
[0033] A total of 39 aptamer sequences were obtained by cloning and sequencing. Using DNAMAN, Mfold software analysis, according to its primary structure, secondary structure, etc., it is divided into 7 families, and representative sequences are selected from each family, and a total of 8 candidate aptamers are selected for characterization analyze. The 8 sequences were marked as HIM-1, HIM-2, HIM-3, HIM-4, HIM-5, HIM-6, HIM-7, HIM-8.
Embodiment 3
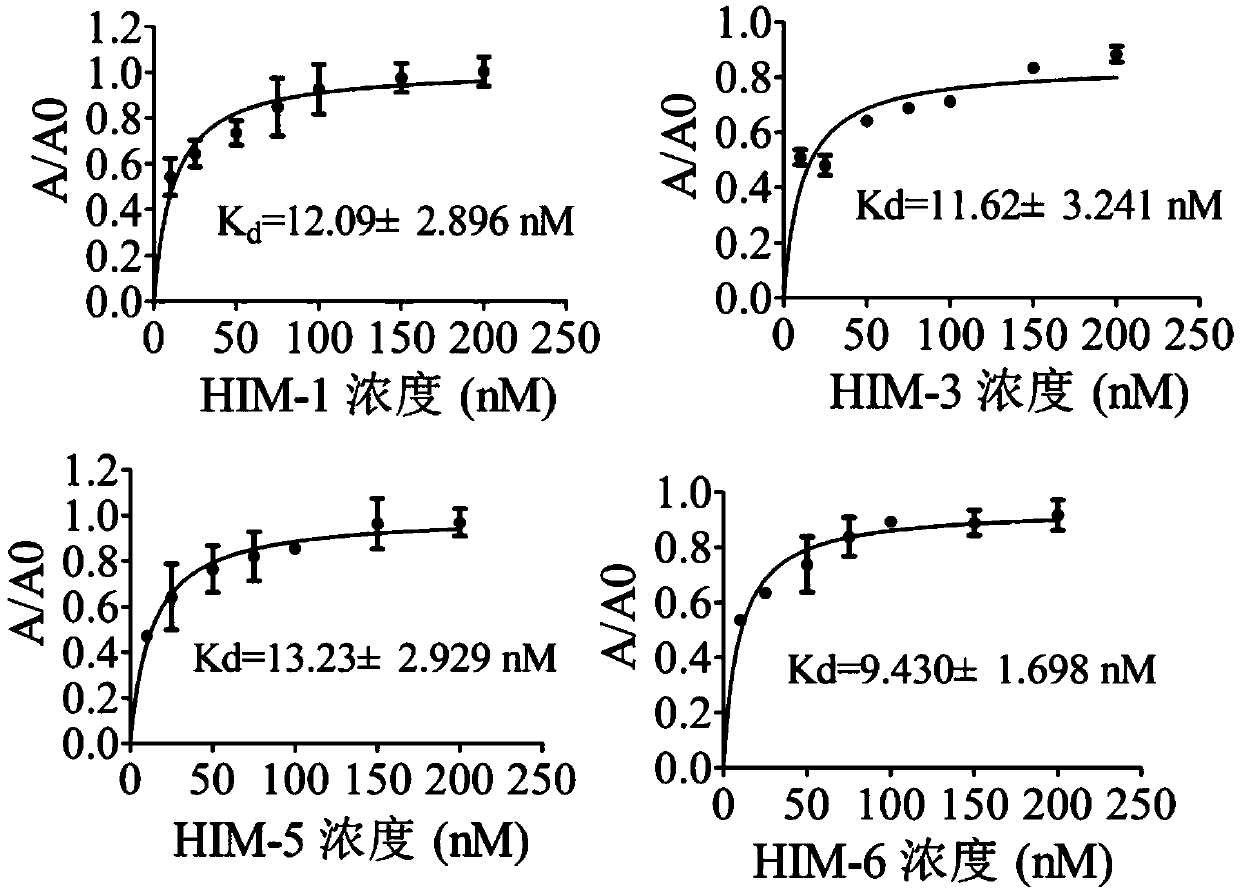
[0034] Example 3: Affinity Analysis
[0035] The above 8 candidate aptamers were synthesized and their 5' ends were biotinylated, diluted with TE buffer (10mM Tris-HCl, 1mM EDTA, pH 7.4) to prepare a 10μM solution, and stored at -20°C for future use. Affinity, specificity, pH stability determination steps are as follows;
[0036] First, add 200 μL of avidin to the microwell plate, and coat overnight at 4°C; wash the plate, add a certain concentration of BSA solution to block at 37°C, 150rpm for 2 hours; wash the plate, biotinylated complementary chain at an appropriate concentration at 37 Bind at 150 rpm for 90 min at ℃; wash the plate and add aptamer to it (experimental group aptamer and target were pre-incubated at 37 °C at 150 rpm for 1 h, blank group with binding buffer (50 mM Tris-HCl, 5 mM KCl , 100mM NaCl, 1mM MgCl 2 , pH7.4) instead of the target) and incubated at 37°C, 150rpm for 90min; wash the plate, and bind avidin-HRP for 1h at 37°C, 150rpm. Wash the plate, add...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com