Quantum dot micelle spherical nucleic acid sensor as well as preparation method and application thereof in Pb < 2 + > detection
A nucleic acid sensor and quantum dot technology, applied in the field of biosensing, can solve problems such as being unsuitable for on-site detection, and achieve the effects of high sensitivity and high specificity detection, good repeatability, and wide detection range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Sequence design of aptamers, DNases, and substrates
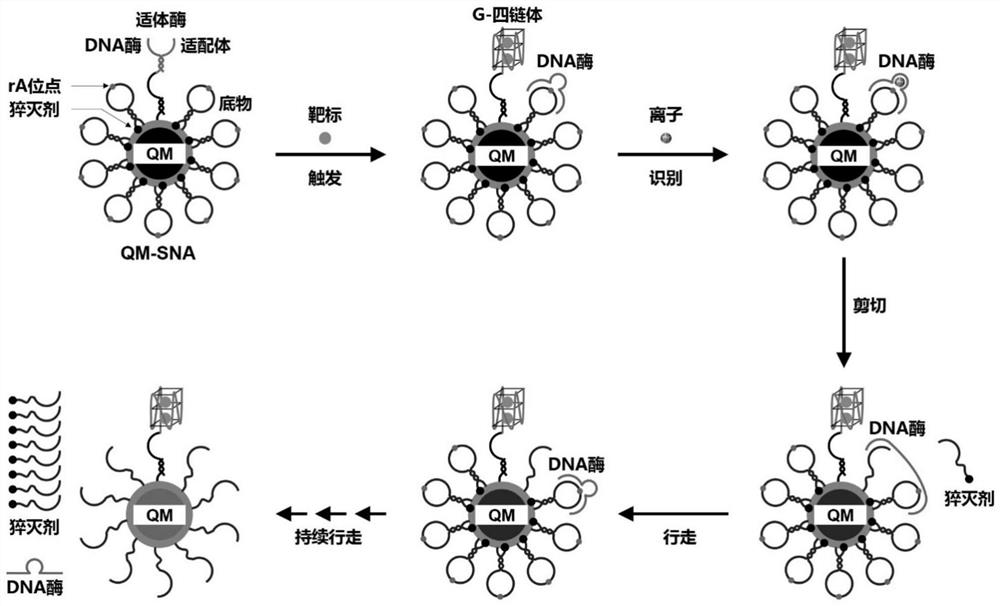
[0035] Such as figure 1 As shown, for the target Pb 2+, and designed the corresponding aptamerase DNA and substrate (Substrate). The aptamyme includes an aptamer sequence and a DNase sequence. The aptamer sequence mainly includes a target binding region and a DNase complementary region, and the amino group at the 5' end is modified for coupling to the QM surface. Among them, the target binding region is the specific recognition sequence of the target, which is used to combine with the target to form a G-quadruplex, and destroy the double-stranded structure of the aptase to release the DNase. The DNase sequence mainly includes three parts: aptamer complementary region, (substrate) orbital binding region, and catalytic center region. Wherein, the aptamer complementary region sequence is the complementary sequence of the aptamer, which is used to hybridize with the aptamer sequence to form aptazyme, immobilize and i...
Embodiment 2
[0040] Preparation of QM-SNA loaded with enzyme and substrate
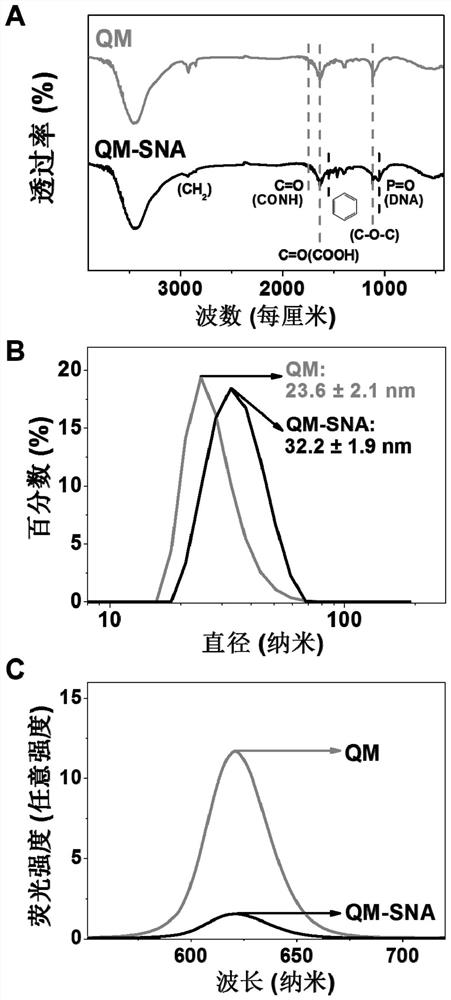
[0041] Quantum dot micelles QM are prepared by ultrasonic film hydration: the quantum dots used are CdSe / ZnS (purchased by Suzhou Xingshuo Nano Technology Co., Ltd.). 0.1 mg of quantum dots and 4 mg of dipalmitoylphosphatidylethanolamine-polyethylene glycol-carboxy (DSPE-PEG2000-COOH) were dissolved in 1 mL of chloroform. A film containing QM was obtained by reacting with a rotary evaporator at 45°C for 15 minutes; 1 mL of deionized water was added to it and dispersed by ultrasonic to prepare a QM solution.
[0042] QM-SNA was prepared by chemical cross-linking method: In a reaction system with a total volume of 500 μL, 10 μL EDC solution (1 mM) was added to 10 nM QM buffer (phosphate buffer solution: 0.01 M, pH 7.4), 10 μL Sulfo- NHS solution (0.5mM), activated at room temperature for 30min. Add 16.7 μL (1 μM) of aptamerase solution and 66.7 μL (1 μM) of enzyme substrate solution to the buffer, stir the reactio...
Embodiment 3
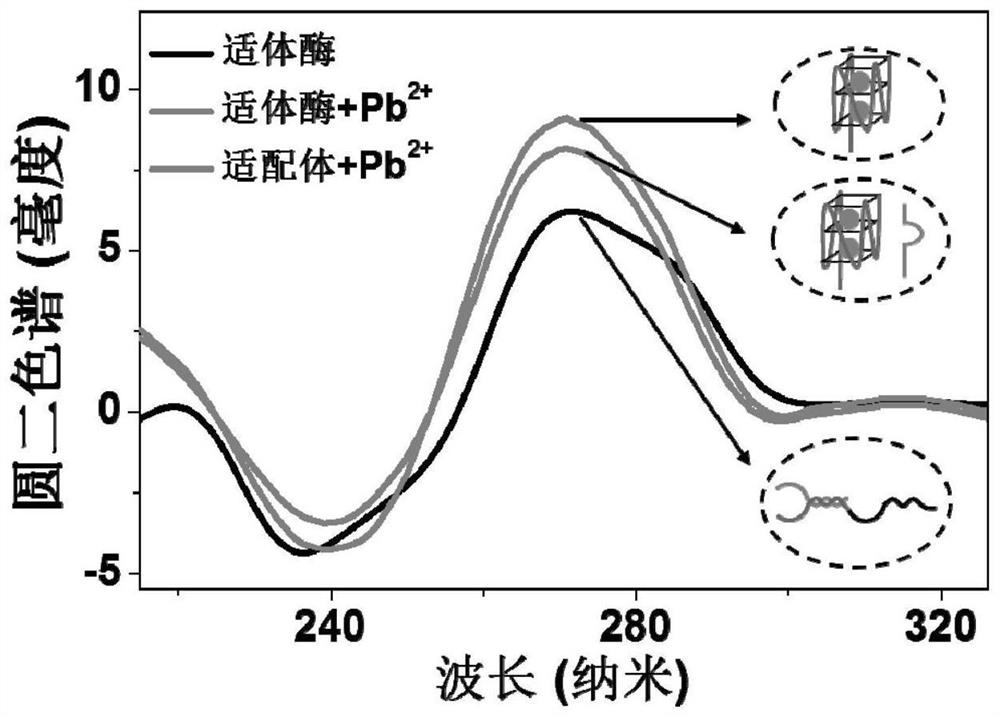
[0045] Pb 2+ triggers a conformational change in aptazymes
[0046] CD spectra of DNA were measured using a Chirascan circular dichroism spectrometer. The aptamase sample was dissolved in 0.01 M pH 7.4 phosphate buffer so that the final concentration was 5 μM, and its CD spectrum was measured. Then add 5000pM Pb to it 2+ , measured its CD spectrum after reacting at room temperature for 30 min. A quartz cell with a 2mm optical path length and an instrument scanner were used to record, the scanning range was 200 to 320nm, and the average value was obtained by scanning three times.
[0047] The result is as image 3 shown, Pb-free 2+ Under these conditions, the aptamerase has a positive peak at 275nm and a peak shoulder at 285nm, showing the characteristics of double-stranded DNA. Join Pb 2+ (5000pM), the aptamerase presents a negative peak at 242nm, and a positive peak at 270nm, which combines with the aptamer to Pb 2+ The typical G-quadruplex formed has the same charact...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com