Pear PbrNSC gene and application thereof
A gene, pear fruit technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve the problems of complex anabolic pathways, unable to meet the needs of high-quality pear fruit for industrial development, and achieve the effect of achieving environmental friendliness and reducing agricultural costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
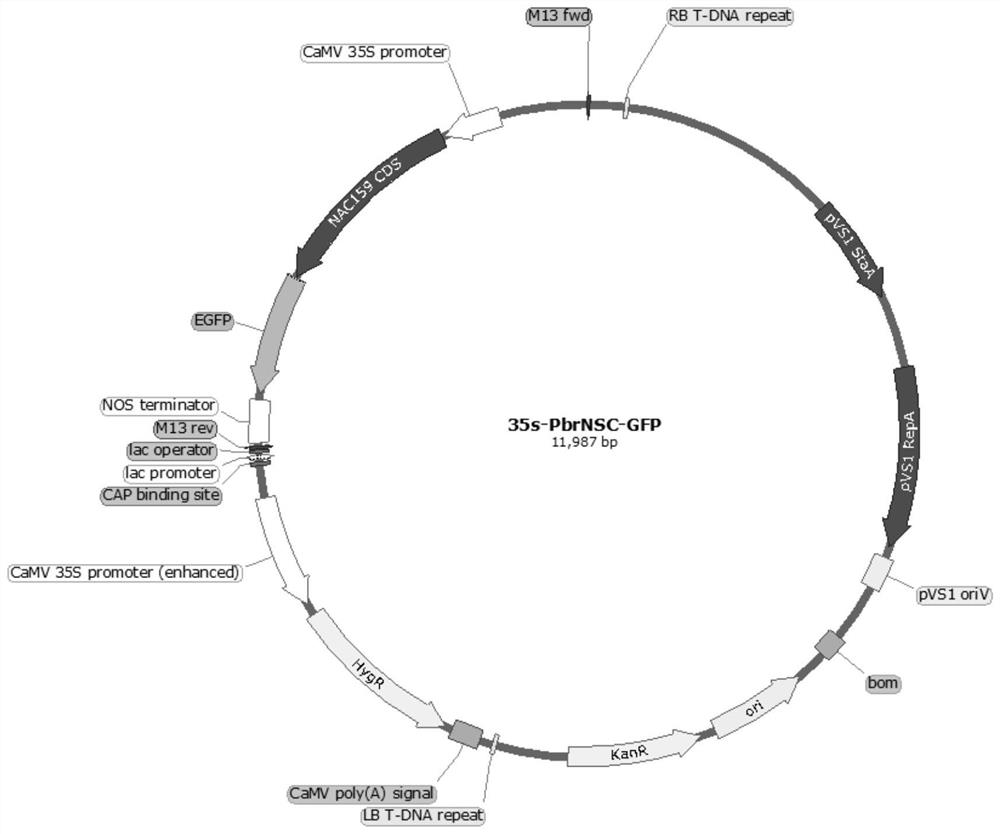
[0025] Example 1 PbrNSC Gene Isolation and Cloning and Construction of Overexpression Vector
[0026] Take 3 μg of early pulp RNA of ‘Dangshan Suli’, and use one-step gDNA removal and cDNA synthesis kit (Transgen, China) for reverse transcription, and the method refers to the instruction manual. According to the analysis of the multiple cloning site of the pCAMBIA-1301 vector and the restriction site on the coding region sequence of the PbrNSC gene, Xba I and BamH I were selected as endonucleases. According to the principle of general primer design, the primers SEQ ID NO.3 and SEQ ID NO.4 with restriction sites were designed with Snapgene software. The 50 μL reaction system included 200ng cDNA, 1× buffer ( GXL Buffer), 0.2μM dNTP, 1.25U GXL polymerase ( GXL DNA Polymerase) (the aforementioned buffer and GXL DNA polymerase were purchased from TaKaRa Company), 0.2 μM of the above primers. The PCR reaction was completed on the eppendorf amplification instrument according to t...
Embodiment 2
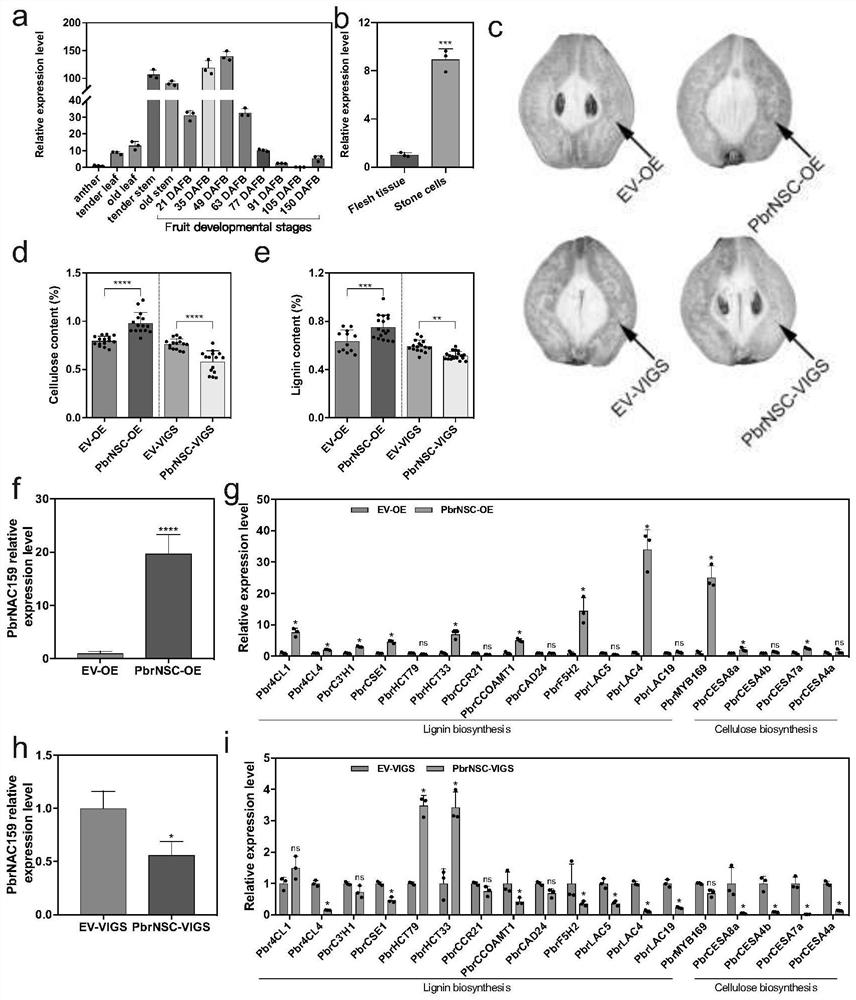
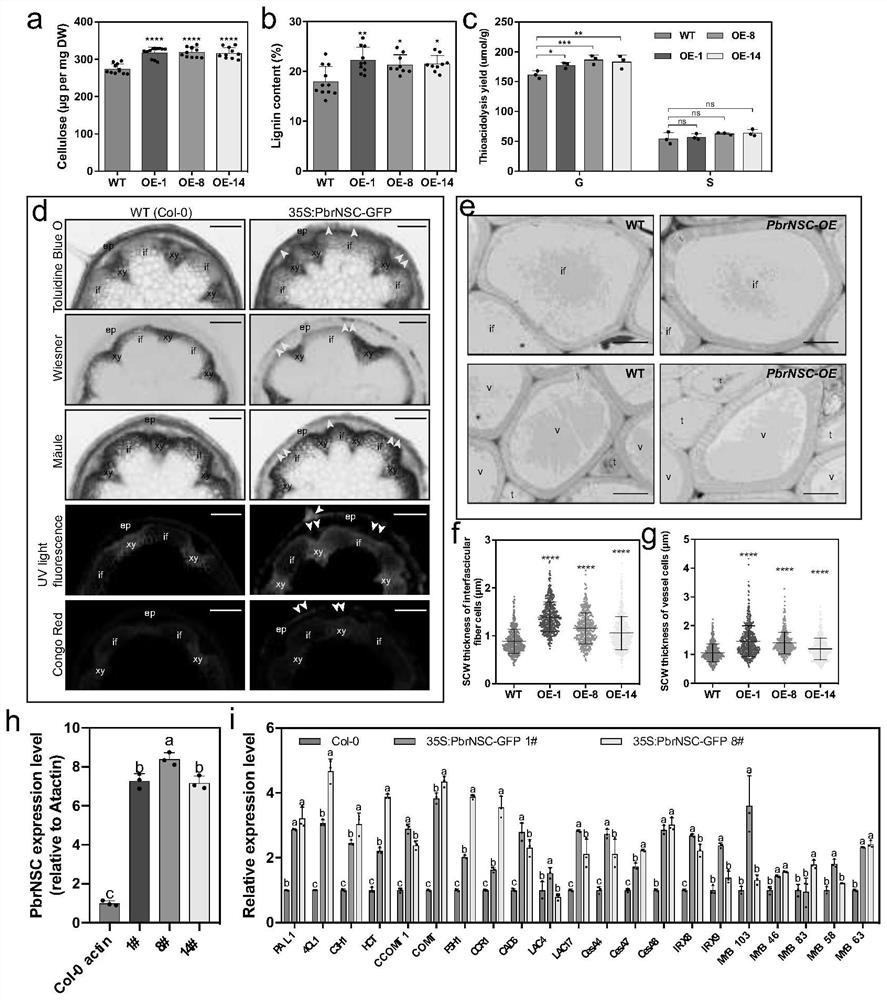
[0028] Example 2 Analysis of spatiotemporal expression pattern of PbrNSC gene
[0029] Different tissue samples of ‘Dangshan Suli’ were collected from Gaoyou Orchard, Jiangsu Province (2015). Total RNA was extracted by CTAB method (Porebski et al., 1997), and the quality of the extracted samples was detected by spectrophotometer and agarose gel. 3 μg of extracted total RNA was used for reverse transcription using the one-step gDNA removal and cDNA synthesis kit (Transgen, China), and the method was referred to the instruction manual. The primers used in the fluorescent quantitative PCR were gene-specific primer pairs: SEQ ID No.5 and SEQ ID No.6; GAPDH was used as an internal reference gene, and the fluorescent quantitative kit was purchased from Roche Company. The instrument used for Real-timePCR was Roche 480 quantitative PCR instrument, and the reaction system was: 10 μL of 2×SYBR GreenI Master Mix, 0.4 μL of upstream and downstream primers (10 μM), 2 μL of cDNA, and 7.2 μ...
Embodiment 3
[0031] Embodiment 3 pear fruit transient transformation
[0032] Use Agrobacterium containing PbrNSC overexpression or gene silencing vector to inject into 'Dangshan Suli' about 35 days after flowering through Agrobacterium-mediated method, the method is as follows:
[0033] 1. Activate the Agrobacterium containing the correct plasmid on solid medium, and grow in an incubator at 28°C for 48 hours;
[0034] 2. Add 30mL containing R to a 100mL Erlenmeyer flask + with K + The liquid LB culture medium, pick the activated Agrobacterium into the activated Agrobacterium with a pipette tip, and grow in a shaker at 28°C and 200rpm for 12 hours;
[0035] 3. Pour the bacterial liquid into a 50mL centrifuge tube and centrifuge at 6000rpm for 15 minutes to collect all the bacterial cells;
[0036] 4. Use an appropriate amount of induction medium (10mM MgCl 2 , 10mM MES, 200mM acetosyringone, pH 5.6) to resuspend the precipitate, adjust the OD value to 0.8-1.2, and induce it with a deco...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com