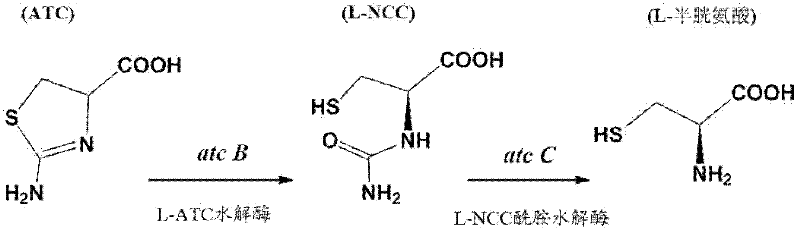
N-carbamyl-L-cysteine (L-NCC) amidohydrolase, encoding gene and application of recombinant expressed protein of L-NCC amidohydrolase
A technology of amidohydrolase and protein, applied in the fields of hydrolase, application, genetic engineering, etc., can solve the problems of few reports on L-cysteine-related enzymes, achieve good industrialization value, high catalytic efficiency, separation The effect of easy purification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Enzyme activity determination method of L-NCC amidohydrolase
[0026] (1) Principle: The acidic ninhydrin method is adopted. L-NCC amidohydrolase decomposes L-NCC to generate L-cysteine, and L-cysteine reacts with acidic ninhydrin reagent under boiling conditions to generate a red substance, which has a maximum absorption at 560nm, and its color depth is the same as The amount of L-cysteine is directly proportional.
[0027] (2) Method: accurately add 6% K to the solution of 1% L-NCC 2 HPO 4 Prepare a solution with a final substrate concentration of 0.75% until the pH value is 7.5, then take 1 mL of the substrate solution, add 0.5 mL of enzyme solution, and react in a 37°C water bath for 10 min.
[0028] The determination of L-cysteine content in the reaction solution adopts the acidic ninhydrin method. Take 0.2mL of the reaction solution, add 0.2mL glacial acetic acid, and then add 0.2mL acidic ninhydrin reagent (weigh 250mg ninhydrin, dissolve in 6mL A mixtu...
Embodiment 2
[0032] Cloning and Primary Structure Characterization of L-NCC Amidohydrolase
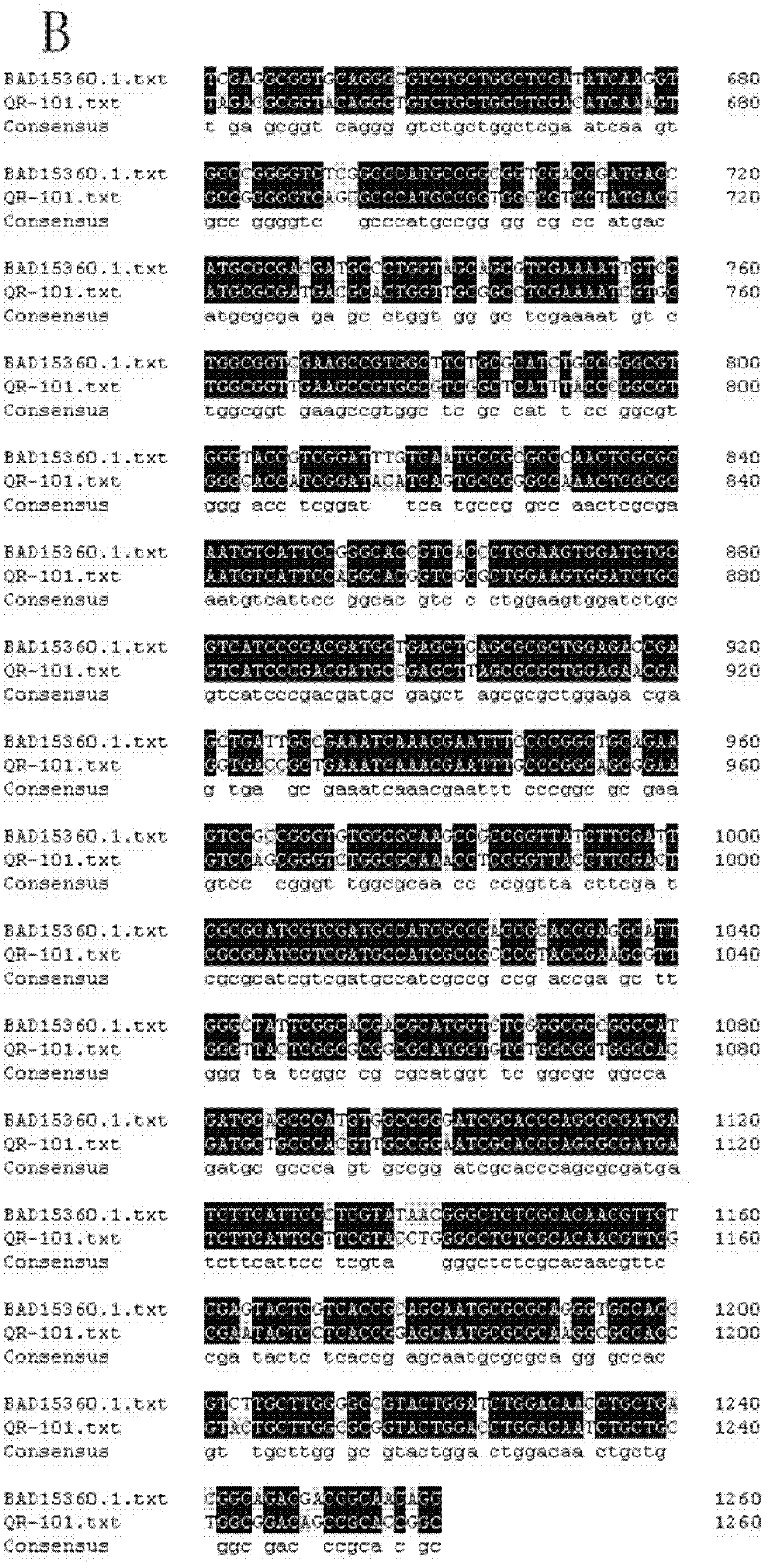
[0033] The present inventor extracted genomic DNA from Pseudomonas sp.QR-101, then digested genomic DNA with HindIII, and recovered 2-9kb HindIII digests from 1.2% agarose gel with a gel recovery kit. Fragments, the recovered enzyme-digested fragments were connected to the Pucl8 vector that had been treated by the same enzyme digestion, and then transformed into E.coli JM109, and blue-white primary screening was performed on a plate coated with X-gal and IPTG.
[0034] The above-mentioned recombinant white single colony containing the insert fragment was picked into each well containing 50 μL LB medium (peptone: 10g / L, yeast powder: 5g / L, sodium chloride: 10g / L), and after overnight culture at 37°C, Add 100 μL of 0.75% L-NCC as a substrate, shake at 35° C. for 30 min, and add acidic ninhydrin reagent for color reaction.
[0035] The recombinants with high activity were re-screened through the micr...
Embodiment 3
[0037] Construction of Escherichia coli Genetic Engineering Bacteria and Expression of Recombinant L-NCC Amidehydrolase
[0038] According to the sequencing results of Example 2, primers P1 and P2 were designed according to the sequences at both ends of the protein-coding gene, wherein upstream P1: 5'-CCG GAA TTC ATG AGT GGA GTC AAC AGC ATG AA-3' contains an EcoRI restriction site; Downstream primer P2: 5'-CCC AAG CTT TCA GCC GGT GCG GCT GTC CGC CA-3' contains a HindIII restriction site.
[0039] Using the genome of strain Pseudomonas sp.QR-101 as a template, DNA amplification was performed according to the following PCR procedure:
[0040] Denaturation at 94°C for 1 min, renaturation at 66°C for 1 min, extension at 72°C for 1 min, and 30 cycles of amplification reaction.
[0041] The PCR amplified product was double digested with EcoR I and HindIII, and connected to the vector pET21a(+) after the same digestion to construct the recombinant pET-21a(+) / atcC.
[0042] The obta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com