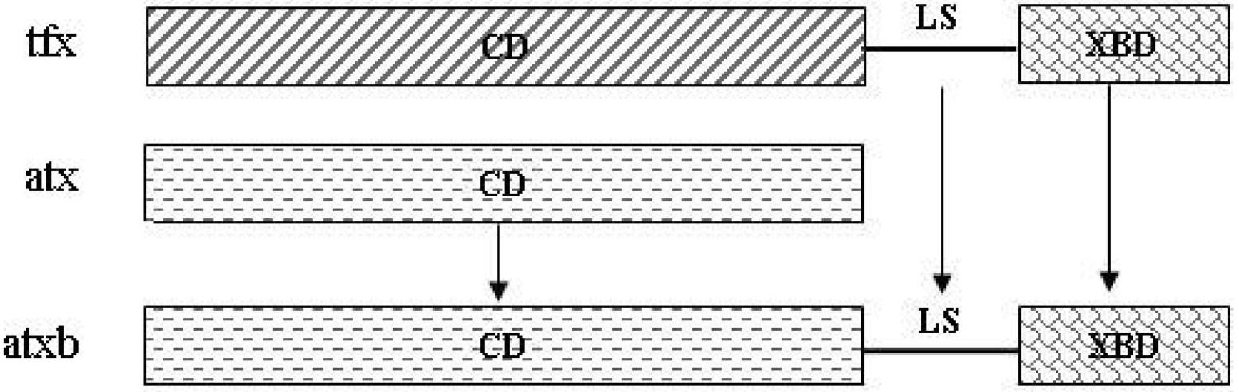
Preparation method of hybrid xylanase atxb
A technology of xylanase and xylan, applied in the direction of microorganism-based methods, biochemical equipment and methods, enzymes, etc., can solve problems such as hindering the hydrolysis of enzyme molecules, and achieve the effect of strong binding ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1: Construction of hybrid xylanase gene atxb
[0053] 1. Preparation of materials:
[0054] Genetic material: hybrid xylanase gene atx and Thermomonospora variegata xylanase A gene, located in pBS-T / atx vector and pBS-T / tfx vector respectively.
[0055] Bacterial species: Escherichia coli TOP10F' was purchased from Invitrogen. Shuttle vector: pPIC9K, purchased from Invitrogen Company, expression host bacteria: Pichia pastoris GS115, purchased from Invitrogen Company.
[0056] High-fidelity DNA polymerase, blunt-ended DNA fragment plus A kit: purchased from Shanghai Sangon Company, and various restriction enzymes and T-vectors were purchased from Promega Company.
[0057] Chemical reagents: Yeast extract and tryptone were purchased from OXOID Company, tissue culture reagents and birch xylan were purchased from Sigma Company. All other chemical reagents were of domestic analytical grade. Primer synthesis: Shanghai Sangon Company or Shanghai Boya Biotechno...
Embodiment 2
[0079] Example 2: Expression of hybrid xylanase gene atxb in Pichia pastoris
[0080] 1. Construction of pPIC9K-atxb
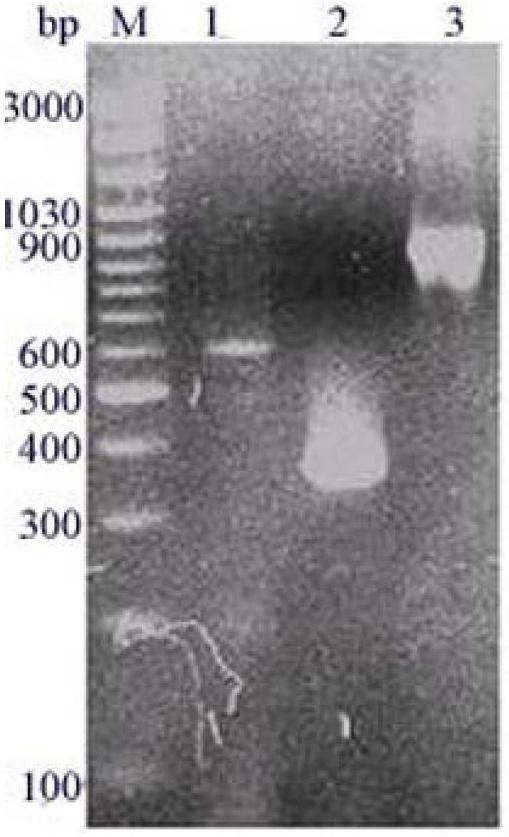
[0081] After sequencing verification, the pBS-T / atxb vector was EcoR 、Not Double enzyme digestion and rubber tapping to recover the target band. The recovered atxb was ligated with pPIC9K, which had also undergone double enzyme digestion, and transformed into Escherichia coli TOP10F'competent cells, picked a single colony on the LB plate containing kanamycin and cultured, and tested by PCR ( Figure 4 ) and double enzyme digestion identification ( Figure 5 ) to obtain a recombinant transformant named TOP10F' / pPIC9K-atxb, and the obtained recombinant expression plasmid was named pPIC9K-atxb.
[0082] 1.1 Double digestion of pPIC9K vector
[0084] Buffer Y + / Tanqo with BSA 10× (purchased from TaKaRa Company) 2.0 μl Eco R (10 U / μl) 1.0 μl not (10 U / μl) 1.0 μl pPIC9K vec...
Embodiment 3
[0145] Example 3: Detection of Yeast Positive Transformants
[0146] 1. Genome extraction of yeast transformants
[0147] Take 1.5 ml yeast transformant liquid, centrifuge at 5000 rpm at 4°C for 5 min, and discard the supernatant;
[0148] Add 500 μl extract solution, and pipette fully to suspend;
[0149] Add 100 μl 10% SDS solution and 300 μl benzyl chloride, shake vigorously;
[0150] Incubate at 50°C for 1 hour, shake and mix once every 10 minutes;
[0151] Add 300 μl 3M NaAC (pH5.2), ice bath for 15 min;
[0152]Centrifuge at 10,000 rpm for 15 min at 4°C, and take the supernatant;
[0153] Add twice the volume of ice-cooled absolute ethanol, and precipitate for 20 min;
[0154] Centrifuge at 10,000 rpm at room temperature for 15 min, discard the supernatant, and wash once with 70% ethanol;
[0155] Naturally dry, add 100 μl TE buffer to dissolve, that is, the genome sample, and store at -20°C.
[0156] 2. PCR detection
[0157] Using the extracted yeast recombinan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com