Cell DNA damage detection method
A technology for DNA damage and detection methods, which is applied in the direction of measuring devices, instruments, and material analysis through electromagnetic means, can solve the problems of sophisticated and complex production processes, high hardware and technical requirements, and achieve simplified preparation processes and improved efficiency , the effect of not easy cell overlap
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] (1) Preparation of splenocyte samples from mice (Cavia porcellus): kill the mice by cervical dislocation, and quickly remove the spleen in an ultra-clean bench after being briefly treated with 75% ethanol, place it in a glass plate, and wash it off with pre-cooled PBS buffer After removing the blood on the surface, immerse it in PBS buffer, and cut it into 1-2mm pieces with ophthalmic surgical scissors 3 of small pieces. Inhale the PBS buffer with the spleen fragments suspended into a disposable sterile syringe (with the needle removed) and press-filter repeatedly, pass through a 300-mesh cell sieve, and collect the cell suspension. At room temperature, centrifuge at 1500rpm for 10min to get the cell pellet, then resuspend it in RMPI-1640, and adjust the cell density to about 8×10 5 individual / mL.
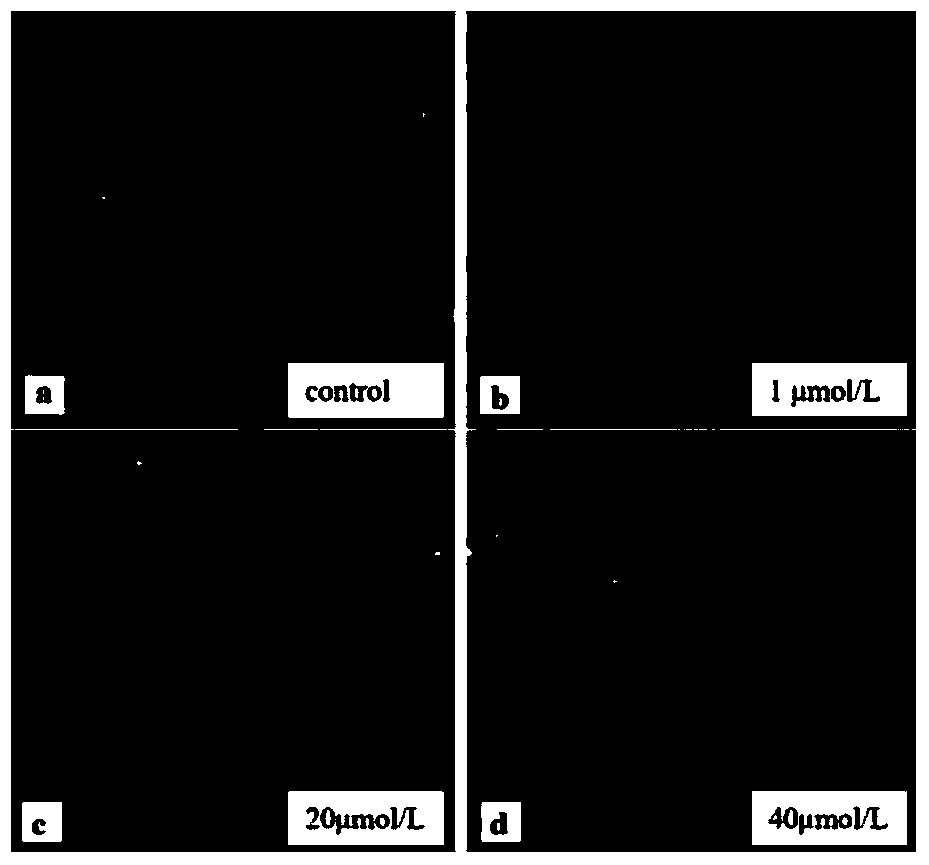
[0036] (2) Cell contamination: seed the cells in a 24-well cell culture plate, and then add H 2 o 2 solution, H 2 o 2 The concentrations were 1 μmol / L, 20 μmol / L, and ...
Embodiment 2
[0046] (1) Preparation of human acute mononuclear cell (THP-1) samples: THP-1 cells were cultured in RMPI1640 medium with 10% fetal bovine serum, 100 U / mL penicillin, and 100 U / mL streptomycin at 37°C for 5 %CO2 cell incubator culture. When passaging, pipette gently with a pipette gun to make the cells uniform and then pass directly. According to the needs of the experiment, the cells are seeded in a 24-well cell culture plate and cultured for 24 hours before use.
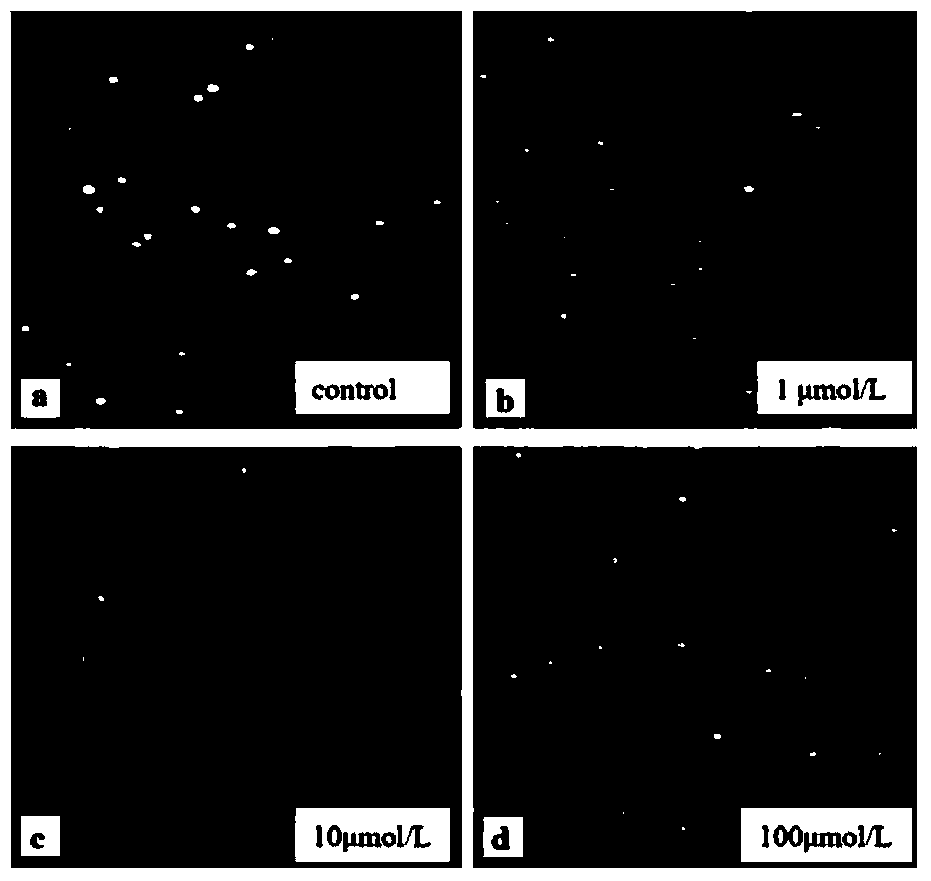
[0047] (2) Cell contamination: seed the cells in a 24-well cell culture plate, then add HgCl 2 solution, HgCl 2 Concentrations were 1 μmol / L, 10 μmol / L, 100 μmol / L, and a blank control group was set up at 4°C for 1.5 hours. After the pollution, wash twice with PBS, collect the cells by centrifugation at 1200rpm, resuspend with PBS, and adjust the cell density to about 8×10 5 pc / mL for use.
[0048] (3) Subsequent operations refer to steps 3-9 of Example 1. See the experimental results figure 2 .
Embodiment 3
[0050] (1) For the preparation of mouse (Cavia porcellus) splenocyte samples, refer to Step 1 of Example 1.
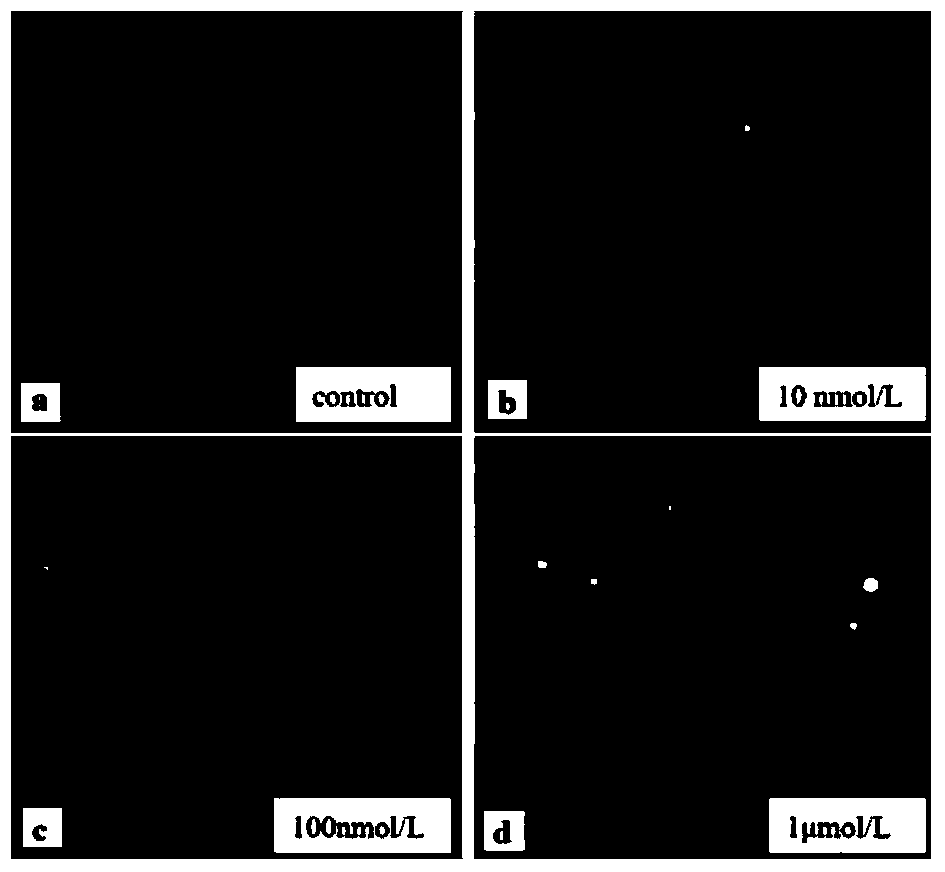
[0051] (2) Cell contamination: seed the cells in a 24-well cell culture plate, then add CuCl 2 solution, CuCl 2 Concentrations were 10nmol / L, 100nmol / L, 1μmol / L respectively, and a blank control group was set up at 4°C for 1.5h. After the pollution, wash twice with PBS, collect the cells by centrifugation at 1000rpm, resuspend with PBS, and adjust the cell density to about 8×10 5 pc / mL for use.
[0052] (3) Subsequent operations refer to steps 3-9 of Example 1. See the experimental results image 3 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com