Gene chip for detecting nine pathogenicity vibrios in marine products
A pathogenic vibrio and gene chip technology, which is applied in the detection/inspection of microorganisms, resistance to vector-borne diseases, DNA/RNA fragments, etc., can solve problems such as complex operations, heavy manual labor, and long inspection cycles. Achieve the effect of strong repeatability, easy operation and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Design and preparation of probes
[0025] 1. Sequence acquisition:
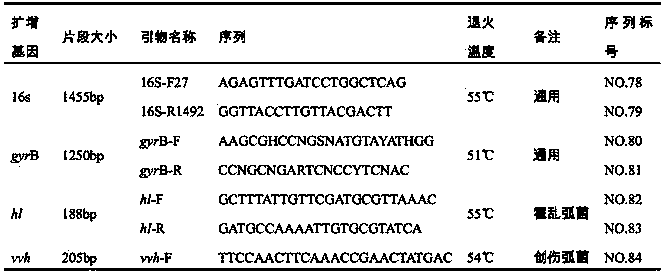
[0026] According to the species ID number of the target species, all the sequence information of the species was searched in the Genbank database, and the genbank format information of all the retrieved sequences was downloaded; the collected related genes are shown in Table 1 below.
[0027]
[0028] 2. Probe design method
[0029] 1) Search the NCBI nucleic acid database to find the gene information of all related species, and download the data in genbank format.
[0030] 2) Extract the gyrb gene sequence from the downloaded data in genbank format.
[0031] 3) For each species of microorganism in the project, use Muscle to perform multiple sequence comparisons of all its gyrb gene sequences.
[0032] 4) According to the multiple sequence alignment results of Muscle, combine the tools to obtain the consensus sequence.
[0033] 5) Take the consensus sequence as input, combine the tool...
Embodiment 2
[0050] Embodiment 2: Design and preparation of strain amplification primers
[0051] of the gene chip of the present invention
[0052] 1. Sequence acquisition: the same as the sequence of the designed probe.
[0053] 2. Primer Design
[0054](1) Design of primers for amplifying interval sequences: After comparing the 16s rRNA of nine Vibrio species downloaded from the public database NCBI with the sequence alignment software Glustal X, a 16s rRNA conserved sequence close to the interval was selected as the upstream primer. And contains Vibrio universal probe sequence.
[0055] (2) Design of primers for amplifying specific gene sequences: compare the specific gene sequences of the above nine species of Vibrio downloaded from the GenBank public database with the sequence comparison software Glustal X, find the conserved segment of the gene, and insert The conserved segment was imported into the primer design software Primer Premier5.0 software. From the output results, sele...
Embodiment 3
[0063] Example 3: Gene Chip Preparation - Chip Spotting
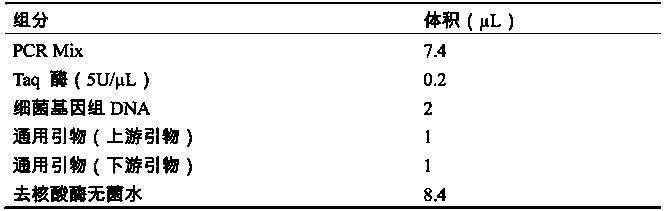
[0064] 1. Dissolve the probes: Dissolve each probe to 40 μM with sterile water, and mix it with the gene chip sample solution at a ratio of 1:1, so that the final concentration of each probe is 20 μM.
[0065] 2. Add plate: Add the dissolved probe to the corresponding position of the 384-well plate, 10 μL per well.
[0066] 3. Spotting: put a clean aldehydized glass slide (Boao Biotechnology) of 57.5mm×25.5mm×1mm (length×width×height) on the stage of the chip spotter (spotarray72), use the control The software (PersonalArrayer16) runs the program, spots in the spotting area on the aldehyded glass slide to form a medium-low density DNA microarray, and the array arrangement rules in the four dot matrix areas on the glass slide are the same.
[0067] 4. Drying: Dry the ordered chips overnight at room temperature, and then dry them in a 45 oven for 2 hours.
[0068] 5. Cross-linking: use a cross-linker to cross-link twice...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com