Gene chip and kit for detecting common pathogenic bacteria in cosmetics
A gene chip and kit technology, which is applied in the field of gene chips and kits for detecting common pathogenic bacteria in cosmetics, can solve the problems of pathogenic bacteria harm, not high throughput, and unable to deal with samples, etc., to achieve strong repeatability, Easy operation and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Probe design and preparation
[0060] 1. Sequence acquisition
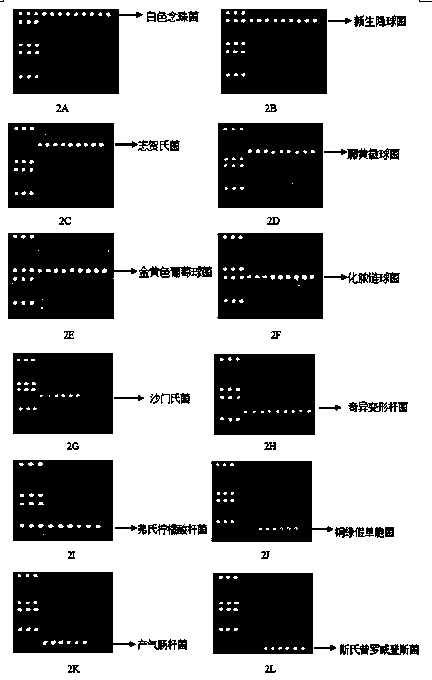
[0061] (1) Acquisition of the 16S-23S rDNA region sequence: Staphylococcus aureus, Streptococcus pyogenes, Salmonella, Proteus mirabilis, Citrobacter freundii, Pseudomonas aeruginosa, Enterobacter aerogenes were downloaded from the GenBank public database , Providencia stutzeri, Shigella and their relatives all 16S-23S rDNA sequences.
[0062] (2) The sequence of the interval region of Micrococcus luteus is not available in the public database. We sequenced its ITS, amplified the ITS of Micrococcus luteus with universal primers for bacterial ITS, and purified the PCR product and connected it to the T vector Above, after electroporation into DH5α competent cells, the plasmids containing 500bp-1kbp were picked and sequenced with ABI 3700 sequencer. The measured sequences were spliced with Staden Package software to obtain the sequence of the M. luteus region.
[0063] (3) Acquisition of the 18S-28S rDNA ...
Embodiment 2
[0076] Gene Chip Preparation: Chip Spotting
[0077] 1. Dissolving probes: Dissolve the probes synthesized in Example 1 in 50% DMSO solution, and dilute so that the final concentration of the probes reaches 1 μg / μl.
[0078] 2. Add plate: Add the dissolved probe to the corresponding position of the 384-well plate, 10 μl per well.
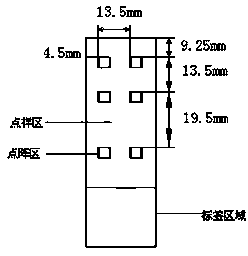
[0079] 3. Spotting: will be as figure 1 The shown 57.5mm×25.5mm×1mm (length×width×height) clean aldehydated glass slide (CEL Associates, Inc.) was placed on the stage of the chip spotter (Spotarray 72), using SpotArray Control software (Tele chem smp3 stealty pin), run the program, press figure 2 The arrangement shown is spotted on the aldehydeized glass slide in the spotting area of 4.5 mm×4.5 mm to form a medium-low density DNA micro-array, and the array arrangement rules in the six dot matrix areas on the glass slide are the same. The size of the dot matrix area is 3mm×2.25mm, the dot pitch in the dot matrix is 250 μm, the matrix: 12×9, ...
Embodiment 3
[0084] Rapid detection of common pathogenic bacteria in cosmetics using gene chips
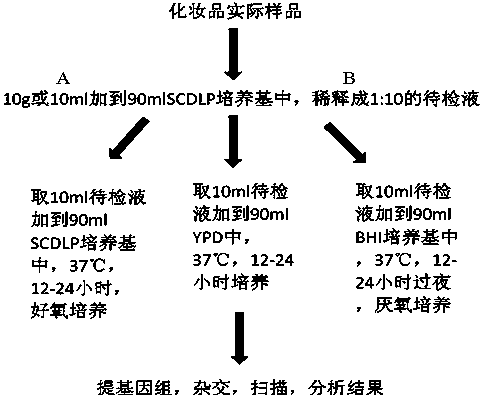
[0085] 1. Sample handling:
[0086] (1) Water-soluble liquid samples: Use a sterilized measuring cylinder to measure 10mL of liquid samples in an ultra-clean bench, add them to 90mL of pre-prepared and sterilized medium, and make a 1:10 dilution. If it is less than 10mL, the dosage can be appropriately reduced, and all samples can be added to the corresponding medium.
[0087] (2) Insoluble oily liquid sample: Measure 10mL of the sample, add it to sterilized mineral oil and mix evenly, then add 10mL Tween-80, mix it in a 42 water bath for about 10 minutes, add sterilized Bacteria pre-enrichment medium 75mL, and then emulsified in a 42°C water bath to make a 1:10 dilution.
[0088] (3) Hydrophilic semi-solid sample: Weigh 10g of sample in a sterile environment, add it to 90mL of sterilized pre-enrichment medium, vortex and oscillate to mix, leave it at room temperature for about 20 minutes, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com