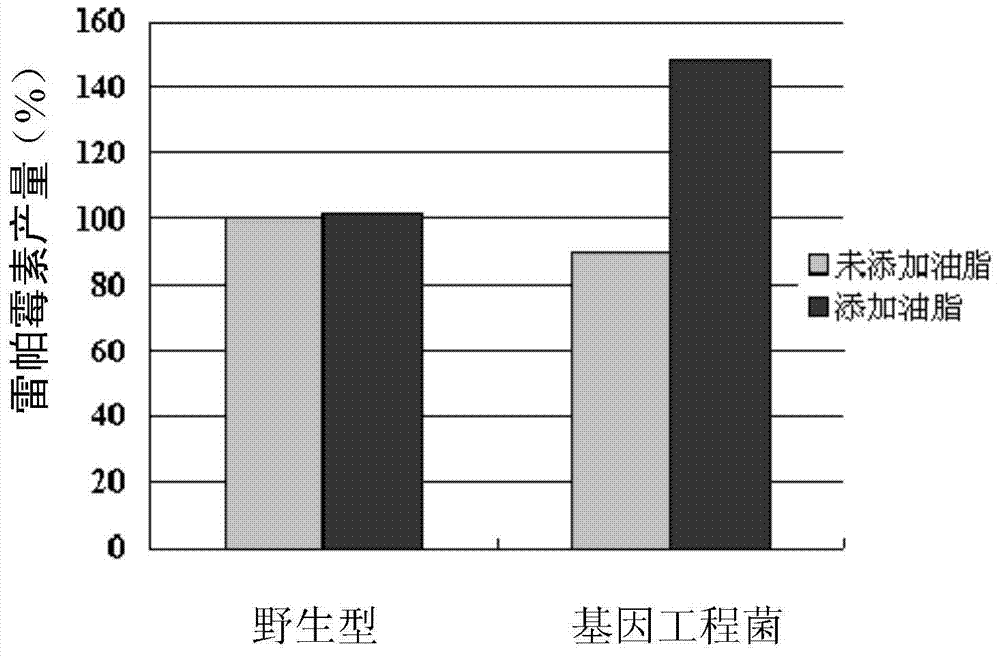
Method for increasing fermentation yield of polyketone compounds
A technology for polyketide compounds and synthase, which is applied in the biological field, can solve the problems of difficulty in meeting practical application requirements and insufficient fermentation yield of polyketide compounds, and achieves the effects of simple and easy operation, low cost and good application prospect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Extraction of Streptomyces roseosporus Genome
[0034] Genome extraction includes the following steps:
[0035] (1) Cultivation of Streptomyces roseospora: inoculate Streptomyces roseospora into 50ml liquid YEME medium, said YEME medium includes: 0.3% yeast extract, 0.5% peptone, 0.3% maltose extract, 1% glucose , 10% sucrose and 5mM MgCl 2 , the stated percentages are mass volume percentages (W / V), cultivated at 28°C (300rpm), so that the bacteria grow to the middle and late logarithmic stages.
[0036] (2) Collection of Streptomyces roseospora: Take 1ml of the culture in a 1.5ml EP tube, centrifuge at room temperature and 8000rpm for 5min, discard the supernatant, and resuspend the pellet in 1ml TE (pH8.0) buffer.
[0037] (3) Phytolysis: add 6 μl of 50 mg / ml lysozyme and react at 37°C for 30 minutes. Add 50 μl of 2mol / L NaCl, 110 μl of 10% SDS, and 3 μl of proteinase K at 20 mg / ml, and act for 3 hours at 50°C or overnight at 37°C (the bacterial liquid sh...
Embodiment 2
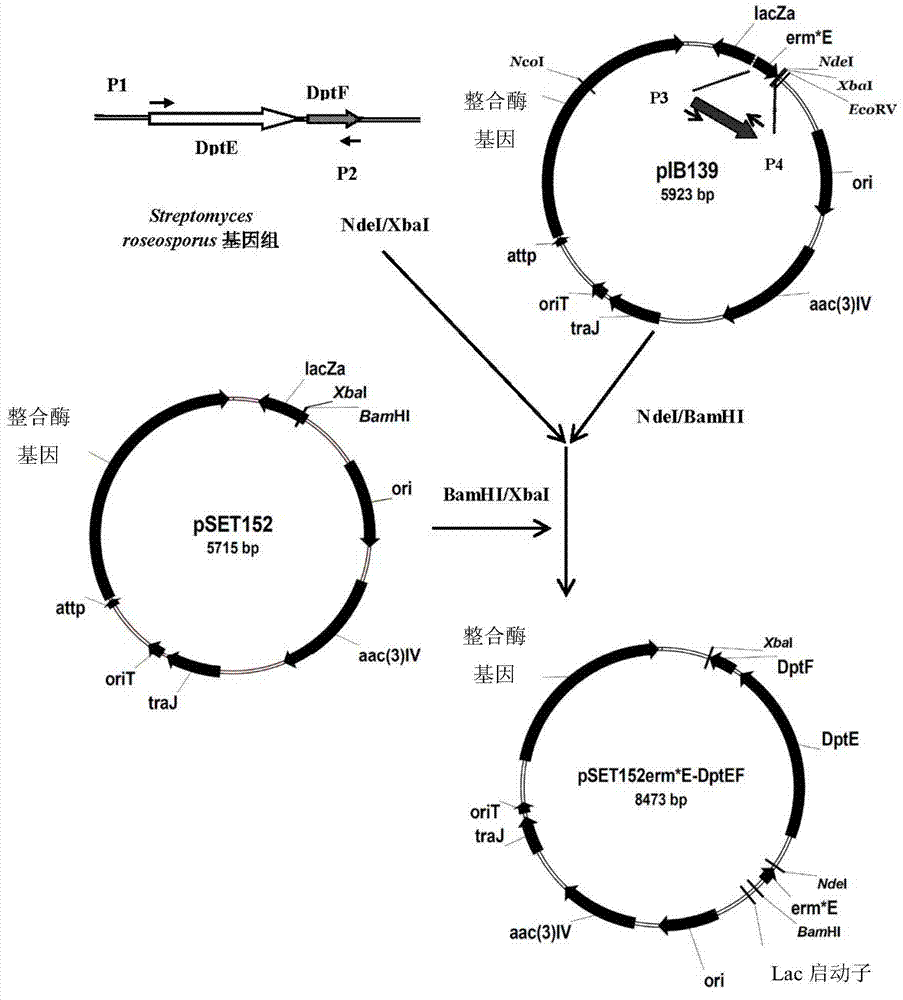
[0042] Example 2 Construction of recombinant expression vector pSET152erm*E-DptEF:
[0043] 1. Cloning of DptEF gene:
[0044] Using the Streptomyces roseospore genome obtained in Example 1 as a template, primers p1 and p2 were used to carry out high-fidelity PCR amplification to obtain a fragment of about 2461bp, which contained complete DptE and DptF genes, and its complete sequence was as SEQ ID NO. 6. Wherein, the sequences of primers p1 and p2 are respectively shown in SEQ ID NO.7 and SEQ ID NO.8.
[0045] (1) The reaction system (50μl) includes: 5μl 10×KOD NEO plus buffer, 3μl 25mM MgCl2, 1.5μl 2.5mmol / L dNTP solution, 2.5μl DMSO, 30pmol P1, 30pmol P2, 1U KOD NEO plus DNA polymerase, 50ng genome , add double distilled water to 50 μl.
[0046] (2) The PCR reaction program is: 95°C for 5min, 30 cycles (94°C for 30s, 56°C for 30s, 72°C for 120s), 72°C for 10min.
[0047] PCR fragments were purified and recovered by agarose gel electrophoresis and gel recovery kit.
[0...
Embodiment 3
[0057] Example 3. Transformation of pSET152erm*E-DptEF into Actinomycetes mobilis:
[0058] Preparation of Actinomycetes spore suspension: About 5 days before the conjugation transfer, take 3 μl of the glycerin preservation solution of Actinomycetes and coat I-Isp2 solid medium, which includes 1% Maltose extract, 0.4% yeast extract yeast extract, 0.4% glucose, 2% agar powder, the stated percentages are mass volume percentages (W / V), and the pH is 7.0. Cultivate at 28°C for about 120 hours, use without Scrape the surface bacteria with a bacteria scraper, wash with 15-20ml of sterile water and shake 10 times on a vortex oscillator through glass beads. After 10 seconds each time, pass the suspension through a sterile syringe with a cotton plug, filter After the liquid is centrifuged at 7000 rpm, the supernatant is discarded, and the spore suspension is obtained by resuspending with an appropriate amount of 0.4-1.0ml sterile water.
[0059] The recombinant expression vector pSET1...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com