DNA molecule for expressing acyltransferase in pichia pastoris and application thereof
A technology of DNA molecules and Pichia pastoris, applied in the direction of transferase, recombinant DNA technology, application, etc., to achieve good biological safety, simple operation, and avoid the effect of endotoxin residues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1. Cloning of acyl homoserine acyltransferase ahlM gene and construction of plasmid pPlCZαA-ahlM.
[0047] 1. Cloning of the acyl homoserine acyltransferase gene ahlM
[0048] The acyl homoserine acyltransferase ahlM gene was found from the Streptomyces sp.M664 strain genome database. The sequence number of the ahlM gene in Genebank is AAT68473.1, and the nucleotide sequence is shown in SEQ ID NO:1 , the amino acid sequence is shown in SEQ ID NO:2. Entrusted Shanghai Yihong Biotechnology Co., Ltd. to synthesize the full nucleotide sequence of the acyl homoserine acyltransferase ahlM gene onto the pUC57 plasmid to obtain the pUC57-ahlM plasmid.
[0049] 2. Construction of pPlCZαA-ahlM recombinant plasmid containing acyl homoserine acyltransferase ahlM gene expressed in Pichia pastoris
[0050] The P1 upstream primer shown in SEQ ID NO:3 and the P2 downstream primer shown in SEQ ID NO:4 were designed according to the nucleotide sequence of the gene encoding acyl...
Embodiment 2
[0058] Embodiment 2, construction and identification of recombinant yeast strain
[0059] 1. For the linearization of the pPICZαA-ahlM recombinant plasmid, the pPICZαA-ahlM recombinant plasmid was digested with SacⅠ restriction endonuclease, and the enzyme digestion system was as follows: the concentration was 100ng / μL pPICZαA-ahlM recombinant plasmid 100μL, SacⅠenzyme 10μL, 30 μL of 10× buffer, 160 μL of water. Enzyme digestion in a water bath at 37°C for 12 hours. After the reaction, add the enzyme digestion system to an equal volume of extract solution consisting of phenol: chloroform: isoamyl alcohol for extraction, wherein the volume ratio of phenol: chloroform: isoamyl alcohol is 25: 24:1. Then centrifuge at 15,000×g for 10min and take the upper aqueous phase, add 2 times the volume of -20°C cold absolute ethanol, mix well, put it into a refrigerator at -20°C for alcohol precipitation for 2h, and then centrifuge again (15,000× g, 10 min), carefully suck off the ethanol...
Embodiment 3
[0071] Example 3, Induced Expression of Recombinant Yeast Strains
[0072] 1. Select 3 clones from the recombinant yeast with the deposit number CGMCC No.10362, inoculate them in 5 mL of YPDS liquid medium containing 100 μg / mL bleomycin, and culture them at 30° C. and 200 rpm / min for 12 hours.
[0073] 2. The culture is inoculated in 20mL BMGY medium according to the inoculum size of 1%, and cultured at 30°C and 200rpm / min for 16-20 hours until OD 600nm Between 2.0-6.0. Centrifuge at 6000rpm for 5min at room temperature, collect the recombinant yeast cells, and then resuspend the cells with BMMY medium to OD 600nm is about 1.0. Continue to culture at 28° C. with shaking at 200 rpm / min for 72 hours, wherein anhydrous methanol is added every 24 hours until the final concentration of methanol is 1%.
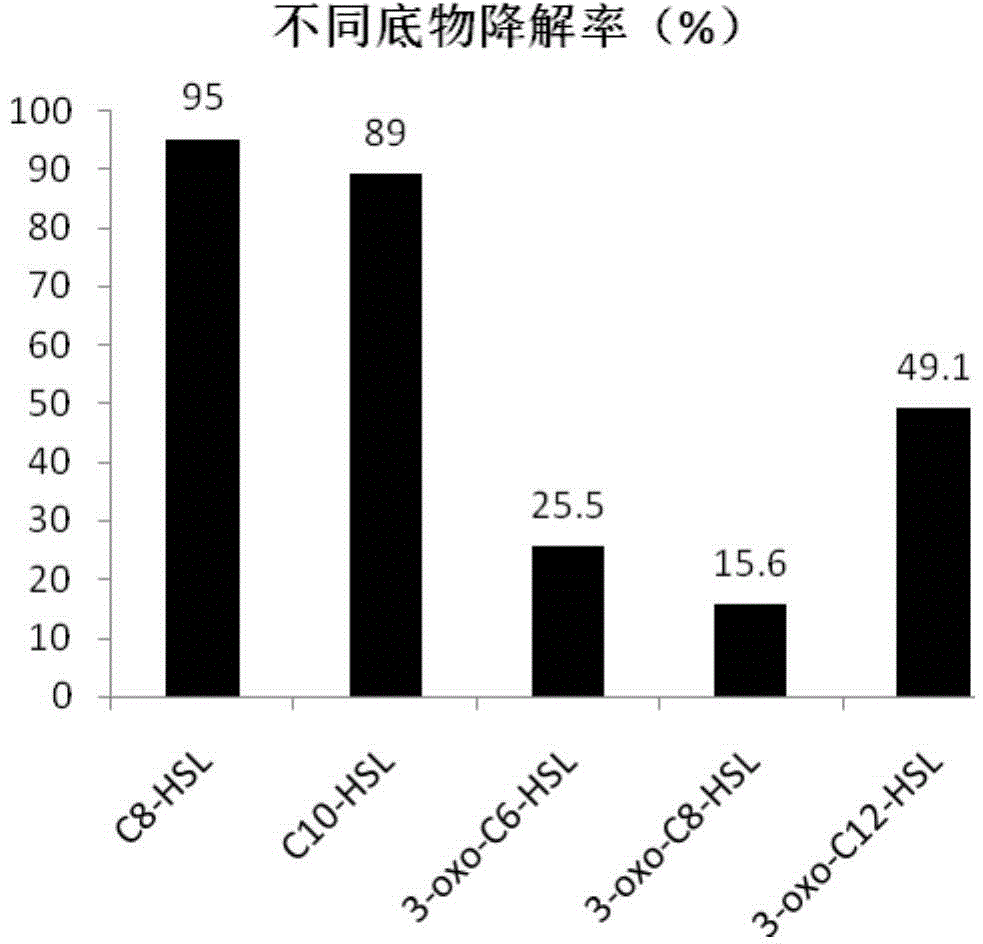
[0074] 3. Take 50 mL of the recombinant yeast liquid induced in step 2 for 72 hours and centrifuge at 5000 rpm / min for 10 min, collect the supernatant after centrifugation, lyoph...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com