ssDNA aptamer for recognizing ofloxacin with specificity and application of ssDNA aptamer
A technology of ofloxacin and aptamer, applied in recombinant DNA technology, DNA/RNA fragments, material testing products, etc., can solve problems such as enhanced phototoxicity, liver cell damage, and drug efficacy reduction. Quick and easy to operate, easy to modify and mark, stable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Construction of random ssDNA library and primers thereof
[0034] (a) Construction of a random ssDNA library with a length of 79 bases
[0035] 5′-TAGGGAATTC GTCGACGGAT CC-N35-CTGCAGGTCG ACGCATGCGC CG-3′ where N represents any one of bases A, T, C, and G, and N35 represents a random fragment length of 35 bases.
[0036] (b) Synthesis of upstream primers
[0037] Upstream primer 1: 5′-TAGGGAATTC GTCGACGGAT-3′
[0038] (c) Synthesis of downstream primers
[0039] Downstream primer 1: 5′-CGGCGCATGC GTCGACCTG-3′
[0040] Downstream primer 2: 5′-biotin-CGGCGCATGC GTCGACCTG-3′
Embodiment 2
[0041] Example 2 In vitro screening of nucleic acid aptamers
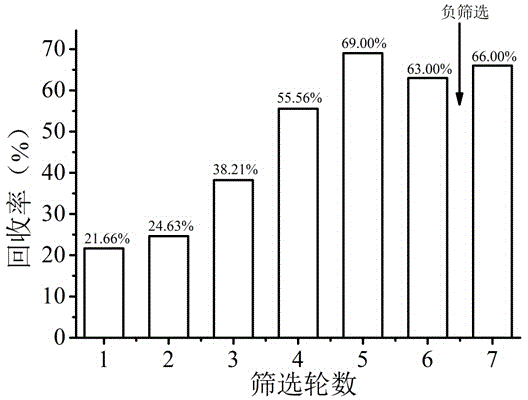
[0042] In order to screen ssDNA aptamers with high affinity and specificity to ofloxacin, a total of 7 rounds of nucleic acid aptamer screening were carried out. In order to improve the specificity of screening, a round of negative screening experiments was carried out after the sixth round of screening. In each round of screening, the recovery rate of ssDNA after the combination of the aptamer and ofloxacin was as follows: figure 1 shown.
[0043] (a) Take 1 μL of 100 μmol L -1 (0.1nmol) of the initial library ssDNA, add 199 μL of binding buffer, mix well, denature at 90°C for 10 minutes, rapidly ice-bath for 10 minutes, place at room temperature for 10 minutes, add the same amount of ofloxacin solution, mix well, and store at room temperature Slightly oscillating for 1 hour, ssDNA self-adaptively forms a three-dimensional structure, and a part of the three-dimensional structure formed by ssDNA can combine with ...
Embodiment 3
[0056] Example 3 Screened ssDNA cloning, sequencing, and structural analysis
[0057] (a) ssDNA clone sequencing
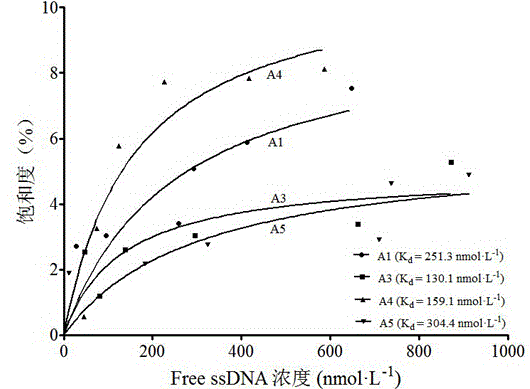
[0058] The ssDNA obtained after the final round of screening was amplified by PCR with upstream primer 1 and downstream primer 1, and the entire amount of the amplified product was loaded on 3% agarose, electrophoresed and the target band was excised, and the ssDNA was recovered by DNA gel recovery kit . Mix 7 μL of the purified PCR product with 1 μL of the pMD-19T vector, ligate overnight at 16°C under the action of T4 ligase, transform into Escherichia coli JM109 competent cells, and culture at 37°C overnight. After colony PCR verification, 21 positive clones were randomly selected and transferred to liquid LB medium, cultured for 12 hours, plasmids were extracted with a plasmid extraction kit, and 12 aptamers of different sequences from A1 to A12 were obtained by sequencing.
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com