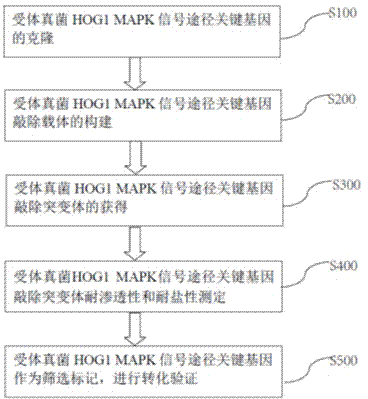
Fungus genetic screening method based on marker gene
A technology of fungi and genes, which is applied in the field of microbial genetic transformation engineering, can solve problems such as the inability to study double genes or multiple genes, and the limited range of use of screening marker genes, and achieve the effect of biological safety and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1: Obtaining the PBS2 gene deletion mutant of Colletotrichum gloeosporioides
[0037] (1) Cloning of the PBS2 gene of Colletotrichum gloeosporioides
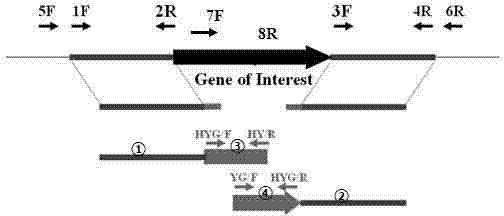
[0038] Search the known fungal HOG1 MAPK signal pathway key gene PBS2 gene sequence in the GenBank database (accession number: NP_012407). Search for homologous genes (Colletotrichum graminicola (M.1.001): GLRG_07565.1; Colletotrichum higginsianum) in the published anthrax genome database (http: / / www.broadinstitute.org / annotation / genome / colletotrichum_group / MultiHome.html) (IMI 349063): CH063_12338.1). Design primer pairs SEQ ID NO:1 and SEQ ID NO:2. The PCR method was used to amplify and clone in Colletotrichum gloeosporioides and sequenced to obtain the PBS2 gene sequence SEQ ID NO: 3.
[0039] Table 1: Sequence information involved in the present invention
[0040] Name
Sequence information
PBS2-OF (SEQ ID NO:1)
CCCAAGCTTATGGCAGACATCAACACCCCC
PBS2-OR (SEQ ID NO: 2)
GCCGAATTC TTCCAACCTGTTCTCCGAGCC
PBS2 g...
Embodiment 2
[0051] Example 2: Determination of permeability and salt tolerance of the PBS2 gene deletion mutant of Colletotrichum gloeosporioides
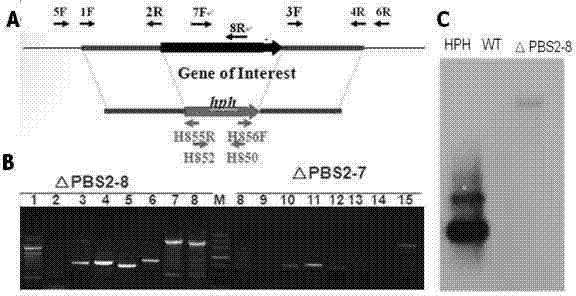
[0052] CgPBS2 gene affects the salt stress function of Colletotrichum gloeosporioides: A comparative analysis of the growth of the wild strain of Colletotrichum gloeosporioides and the gene knockout mutant △PBS2-8 on the medium containing high concentration of NaCl. The results showed that the gene deletion mutant grew slowly in a medium containing high concentration of salt (see Figure 4 ), indicating that the PBS2 gene deletion mutant is sensitive under high salt conditions. It is confirmed that high-salt medium (PDA+1M NaCl) can be used as a screening medium.
[0053] CgPBS2 gene regulates the function of Colletotrichum gloeosporioides against osmotic pressure: Comparative analysis of the wild strain of Colletotrichum gloeosporium and the gene knockout mutant △PBS2-8 grow on the medium containing glucose, sucrose and sorbitol. Gene deletion mu...
Embodiment 3
[0054] Example 3: Using △PBS2-8 as the recipient bacteria and PBS2 gene as the selection marker gene, GFP gene was transformed.
[0055] First, the pSK-PBS2-GFP expression vector needs to be constructed. Using pBluescript Sk+ as the vector backbone, use SalI / ClaI and BamHI / XbaI to connect the promoter (from pCSN43 (C.Staben, 1989)) and terminator (from pAN7-1) of the gene trpC, and then use the primer pair GFP -F / GFP-R was amplified from the vector pSULF·gfp to obtain the PCR product fragment of eGFP, which was connected with PstI / BamHI double enzyme digestion ligation method to obtain pSK-G (see figure 2 ). The primer pair PBS2-OF / PBS2-OR was used to amplify the ORF sequence of Colletotrichum gloeosporioides PBS2 gene (without terminator), and it was connected to pSK-G by Hind3 / EcoRI double enzyme digestion method to obtain pSK-CgPBS2-GFP.
[0056] As in Example 1, using PEG-mediated transformation of protoplasts, the plasmid pSK-CgPBS2-GFPLF was introduced into the △PBS2-8 gene...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com