Phosphatidase, coding gene, preparation method and application thereof
A phospholipase and coding technology, applied in the fields of coding genes, phospholipase, and preparation, can solve the problems of low temperature stability, affecting the quality of finished products, temperature stability of residual phospholipase, etc., achieving broad pH stability, broad application prospects, The effect of excellent thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
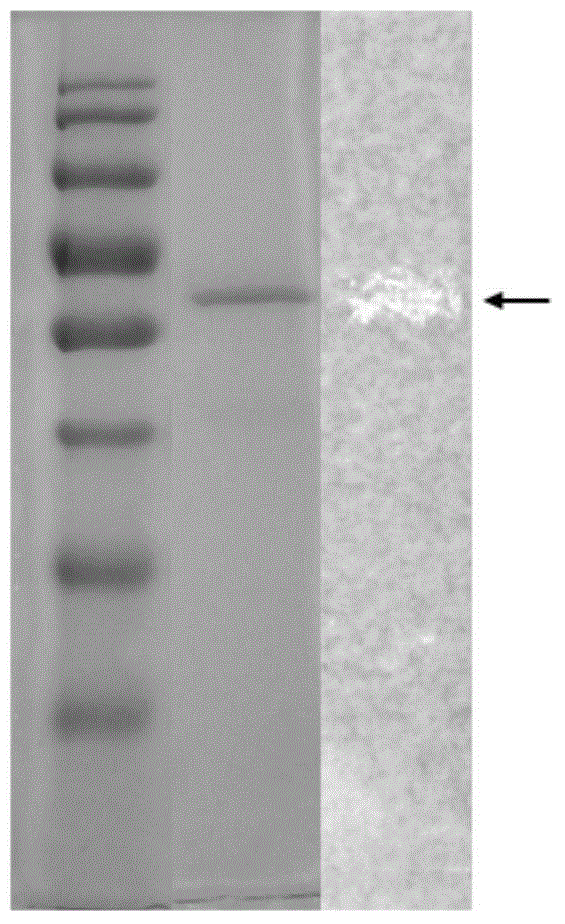
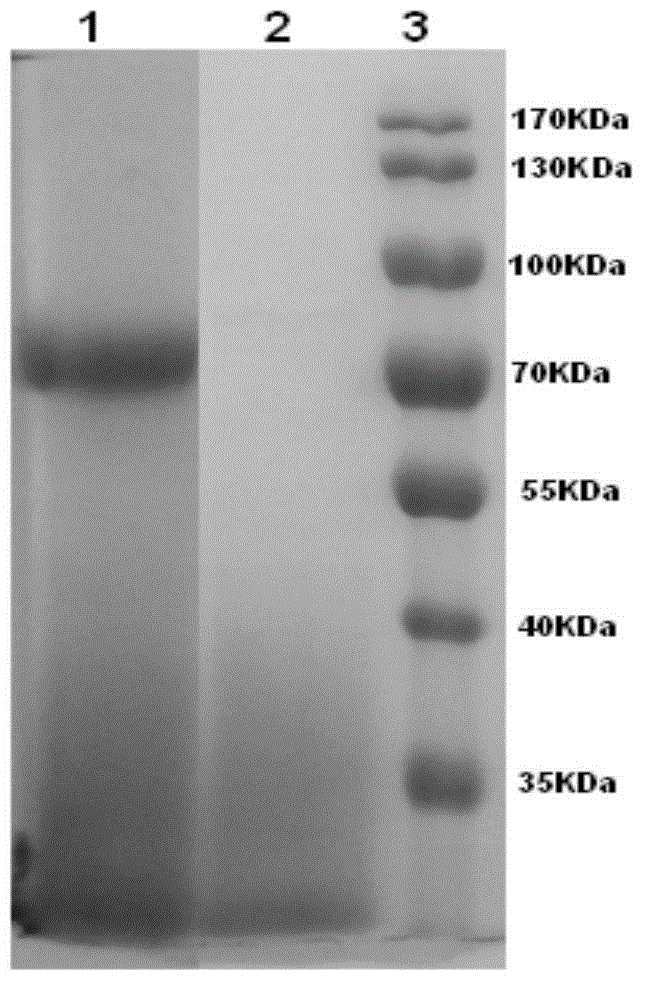
[0137] Example 1: Separation and purification of wild polypeptide enzyme of the present invention and identification of partial amino acid sequence information
[0138]Take 2 freshly cultured Cladosporium WBRD00050 (CGMCC No.7508) slopes (incubate on PDA medium at 28°C for 120 h), add 10 ml of sterile water to each slope, wash thoroughly to obtain a spore suspension, and combine all the spore suspensions liquid. Then inoculate the spore suspension into the fermentation shaker flask according to the inoculum amount of 1ml / bottle. The fermentation shaker flask has a specification of 250ml, and the filling volume is 50ml. The culture conditions of the fermentation shake flask are: 28° C., 200 rpm, and the culture time is 120 hours.
[0139] The composition of the fermentation medium is as follows:
[0140] Sucrose 1%, peptone 0.5%, yeast extract powder 0.5%, corn steep liquor 0.5%, Na 2 HPO 4 0.25%, KH 2 PO 4 0.15%, magnesium sulfate 0.1%, zinc sulfate heptahydrate 0.2%, a...
Embodiment 2
[0164] Embodiment 2: the cloning of polypeptide coding polynucleotide sequence of the present invention
[0165] The total RNA of Cladosporium WBRD00050 was extracted according to the instructions of Qiagen RNeasy Plant Mini Kit. According to the instructions of Promega Reverse Transcription Kit, RNA was reverse transcribed into cDNA. Then, degenerate primers were designed according to the amino acid sequence information of the three peptides obtained in Example 1. SEQ ID NO:3 is an upstream primer designed based on the sequence of PEAEY in peptide 1, and its degeneracy is 128. SEQ ID: 4 is an upstream primer designed based on the YQPVE sequence in peptide 2, and its degeneracy is 128. SEQ ID NO:5 is an upstream primer designed based on the GAGFD sequence in peptide 3, and its degeneracy is 256.
[0166] Using SEQ ID NO: 3-5 as an upstream primer, oligo dT as a downstream primer for PCR, and using the cDNA prepared above as a template to perform PCR. The PCR conditions are...
Embodiment 3
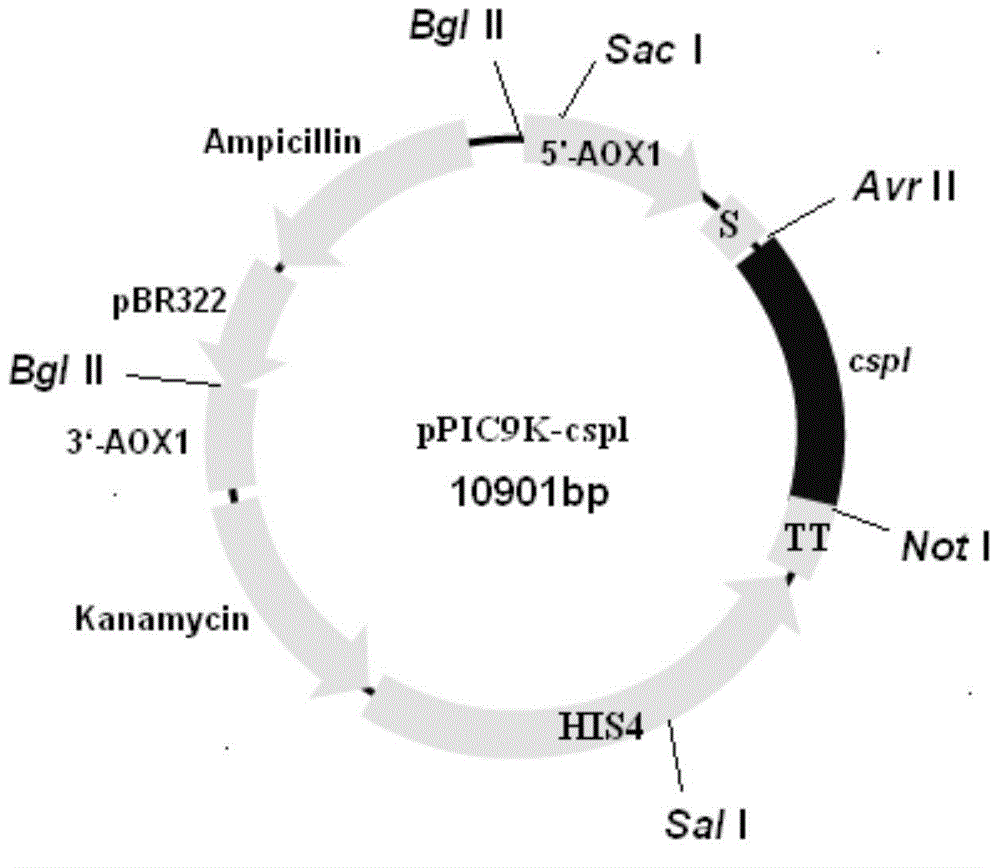
[0169] Embodiment 3: Construction of polypeptide expression vector and engineering bacteria of the present invention
[0170] SignalP 4.1server software (http: / / www.cbs.dtu.dk / services / SignalP / ) analyzed the polypeptide shown in SEQ ID NO: 2, and as a result, the amino acid sequence at positions 1-20 was determined to be the signal peptide sequence. Then, based on the sequence without the signal peptide, the primers were designed. The upstream primer is shown in SEQ ID NO: 8, and the downstream primer is SEQ ID NO: 9.
[0171] Using the cDNA prepared in Example 2 as a template, the upstream and downstream primers shown in SEQ ID NO:8 and SEQ ID NO:9 were used as primers to clone the target sequence.
[0172] Use 1% agarose gel electrophoresis to separate and purify the target band, and use the E.Z.N.ATM gel recovery kit from OmegaBio-Tek, USA to recover the target band. The products recovered from the gel were digested with two restriction endonucleases Avr II and Not I, and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com