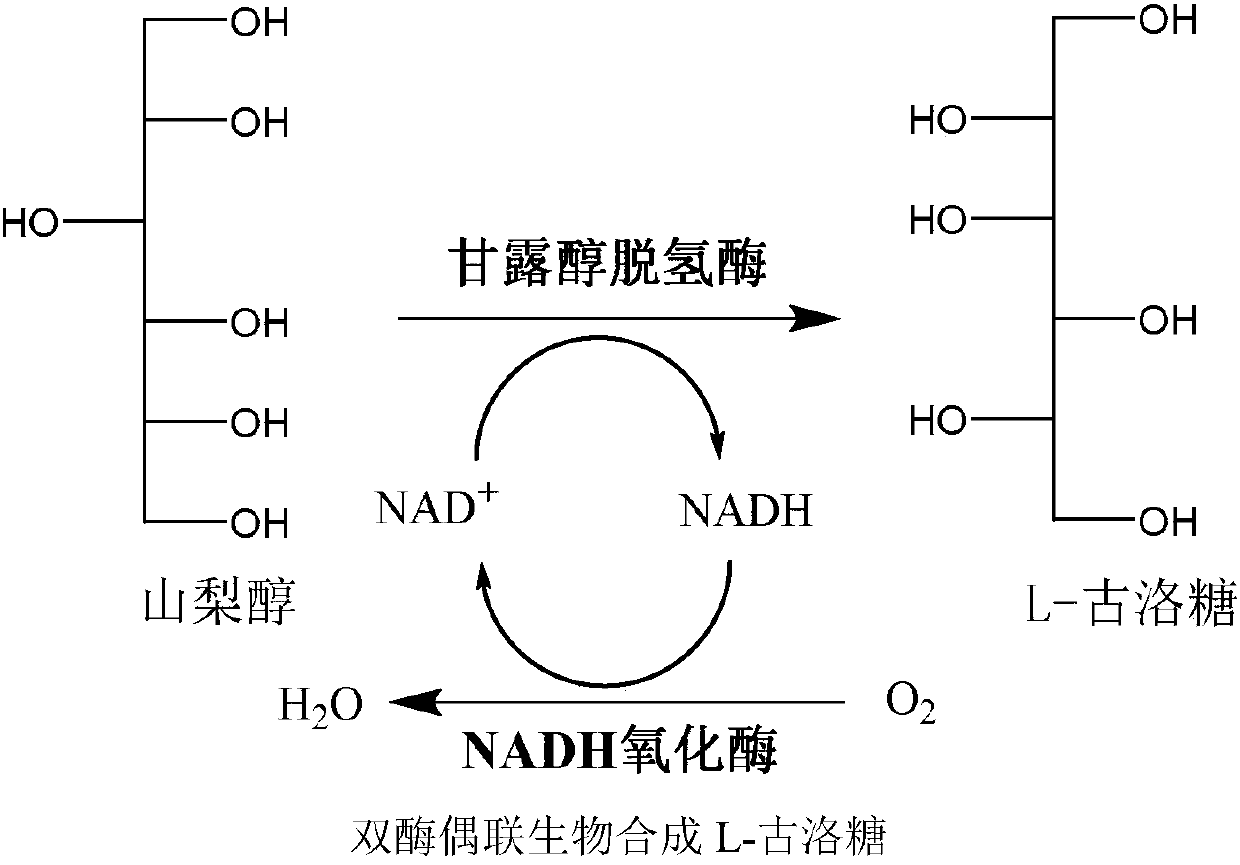
Preparation method for L-gulose
A technology of gulose and oxidase, which is applied in the field of rare sugar biosynthesis, can solve the problem of low production efficiency of L-gulose, and achieve the effect of high-efficiency green biosynthesis, advanced technology, and easy industrialization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
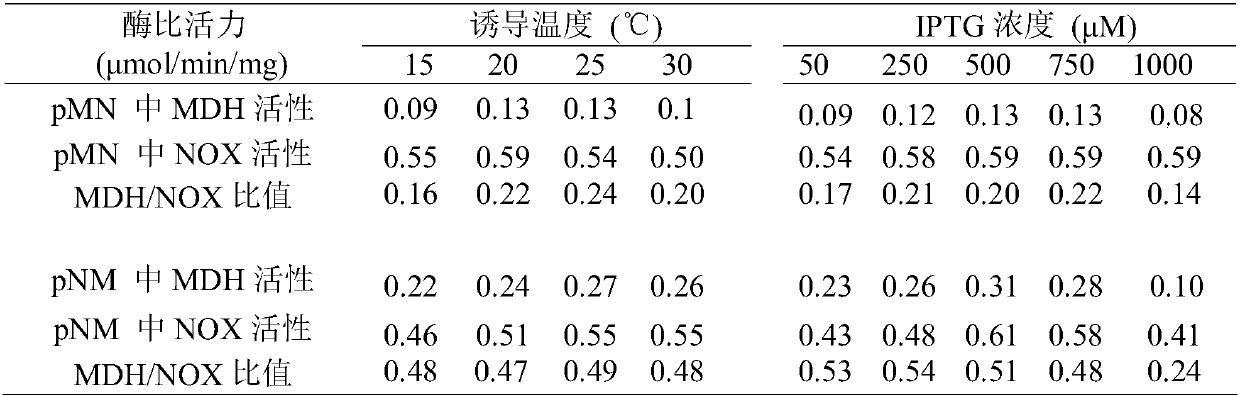
[0018] Embodiment 1 Mannitol dehydrogenase and NADH oxidase activity assay method
[0019] Mannitol dehydrogenase activity assay: 0.6 μmol NAD in 1 mL reaction system + , 0.60 μmol of D-sorbitol, 0.1M Tris-HCL (pH 8.0) and 10 μl of crude enzyme solution. The absorbance change of the reaction system at a wavelength of 340 nm was detected within 1 min at 32°C. One mannitol dehydrogenase enzyme activity unit is defined as the number of micromoles of NADH produced per minute at 32°C, and the specific activity of the enzyme is defined as the number of activity units per mg of total protein.
[0020] Determination of NADH oxidase activity: 0.6 μmol NADH, 0.1M Tris-HCL (pH 8.0) (air saturated) and 10 μl crude enzyme solution were contained in 1 mL reaction system. The absorbance change of the reaction system at a wavelength of 340 nm was detected within 1 min at 32°C. One NADH oxidase enzyme activity unit is defined as the number of micromoles of NADH consumed per minute at 32°C, ...
Embodiment 2
[0021] Example 2 HPLC detection method of L-gulose (PMP derivatization method)
[0022] Take 50 μL of L-gulose standard solution or diluted reaction solution and place it in a centrifuge tube, add an equal volume of 0.6M sodium hydroxide solution, add 100 μL of 0.5M PMP methanol solution, and react at 70°C for half an hour. Take it out and let it cool naturally, add 50 μL of 0.6M hydrochloric acid to neutralize the reaction, add 1 mL of sterile water to mix and dilute. Add 0.8 mL of chloroform, shake and mix well, centrifuge at 12,000 rpm, take the water layer, filter with a 0.22 μm filter membrane, and perform HPLC analysis.
[0023] HPLC analysis method:
[0024] Chromatographic column: YMC C18 column, 150mm×4.6mm, 5μm
[0025] Mobile phase: A: ultrapure water (containing 0.1% formic acid) B: acetonitrile (containing 0.1% formic acid)
[0026] Gradient: 20% B isocratic elution for 20min
[0027] Column temperature: 40°C; Flow rate: 1ml / min; Wavelength: 245nm;
[0028] I...
Embodiment 3
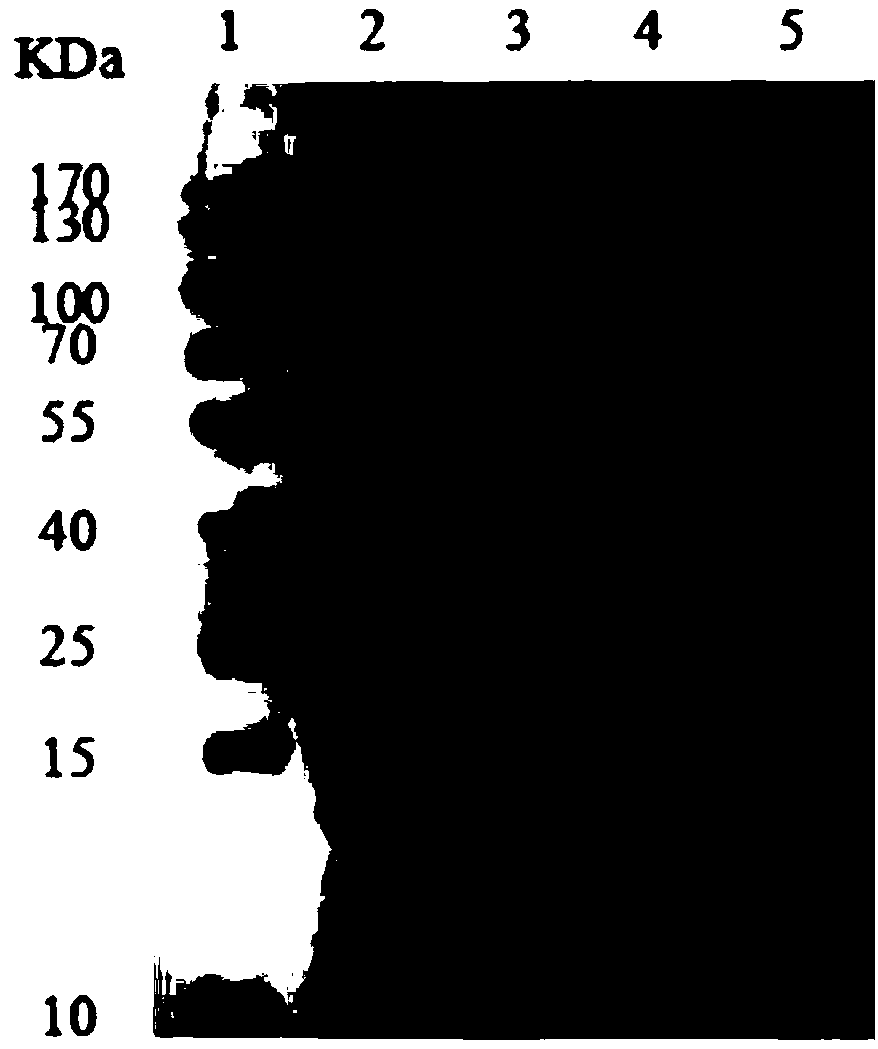
[0030]Example 3 Construction of heterologous expression strains of mannitol dehydrogenase and NADH oxidase
[0031] Use Nco I and EcoR I to insert the mannitol dehydrogenase gene mdh into the multiple cloning site I of pACYCDuet-1, and Nde I and Xho I to insert the NADH oxidase gene nox into the multiple cloning site II to obtain the recombinant plasmid pACYCDuet-mdh- nox. Similarly, use Nco I and EcoR I to insert nox into multiple cloning site I, and use Nde I and Xho I to insert mdh into multiple cloning site II to obtain pACYCDuet-nox-mdh. On this basis, the two plasmids were transformed into Escherichia coli BL 21 to obtain recombinant expression strains E.coli (pACYCDuet-mdh-nox) (pMN for short) and E.coli (pACYCDuet-nox-mdh) (pNM ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com