Nucleic acid sequence for detecting diabetes risk site, kit and detection method thereof
A nucleic acid sequence and nucleotide sequence technology, applied in the field of nucleic acid sequence detection of diabetes risk sites, can solve the problems of restricted development and application, easy pollution, complicated operation, etc., to avoid non-specific amplification problems, and to achieve a high degree of automation , the effect of simplifying the operation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1: Design of primers and verification templates for 5 polymorphic sites in diabetes
[0045] The ARMS primers selected in the present invention are designed for five polymorphic sites of diabetes (rs228648, rs114202595, rs7754840, rs7756992 and rs7574865). The selected ARMS primers are a set of primer combinations artificially added with mismatched mutation sites at the 3' end of a sequence and its vicinity. By screening primers capable of distinguishing alleles at a single site, the accuracy of PCR amplification is ensured. Highly specific.
[0046] After experimental verification, the specific detection primers for the 5 polymorphic sites of diabetes were finally determined, and the nucleotide sequences were as follows:
[0047] UST2 gene rs228648 site wild-type upstream primer (DM1-F1), mutant upstream primer (DM1-F2) and common downstream primer (DM1-R1)
[0048] DM1-F1 (SEQ ID NO: 1): 5`-tgctcggactcataaatcctt-3`
[0049] DM1-F2 (SEQ ID NO:2): 5`-tgctcgga...
Embodiment 2
[0093] Embodiment 2: Preparation of 5 polymorphism kits of diabetes
[0094] For the five polymorphic sites, the wild-type upstream primers, mutant upstream primers and common downstream primers designed in Example 1 were used for synthesis, and SYBR Green I dye was used for staining, which is a dye that binds to dsDNA The dye with green excitation wavelength in the minor groove region of the double helix, in the free state, SYBR Green I emits weak fluorescence, but once combined with double-stranded DNA, the fluorescence is greatly enhanced, so the fluorescence intensity of SYBR Green I is related to the amount of double-stranded DNA Correlation, so the amount of double-stranded DNA present in the PCR system can be detected based on the fluorescent signal.
[0095] A kit for detecting diabetes risk loci, including 10 PCR reaction solutions, positive controls, negative controls, and chromogenic reagents.
[0096] There are 10 kinds of PCR reaction liquids among the present in...
Embodiment 3
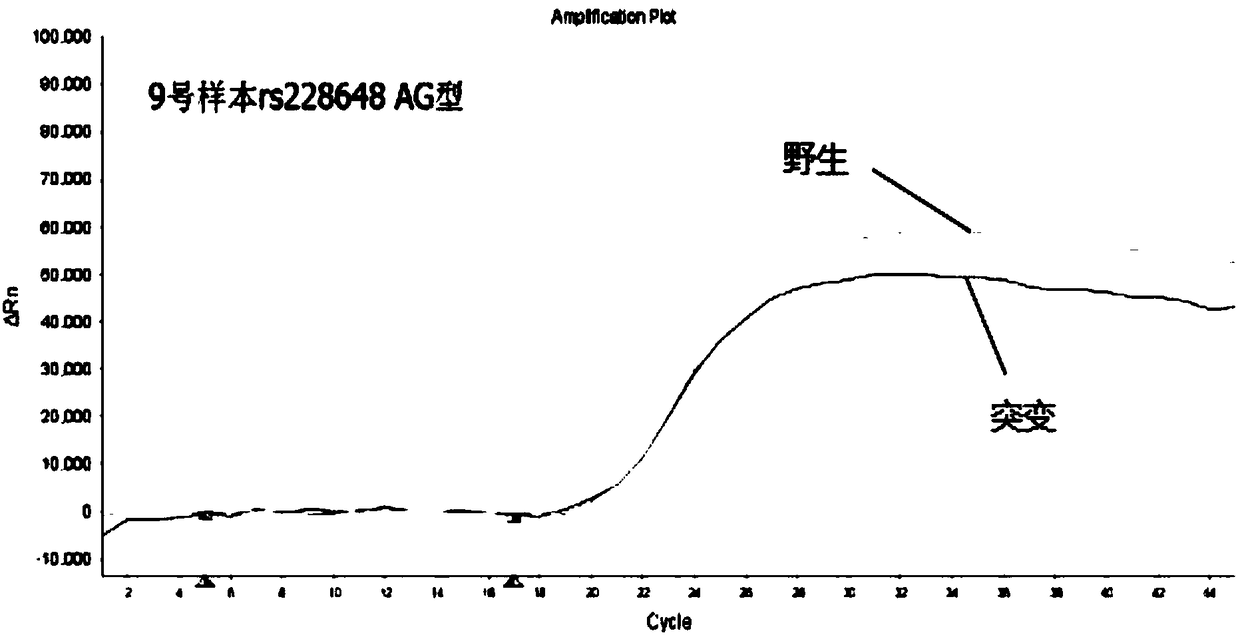
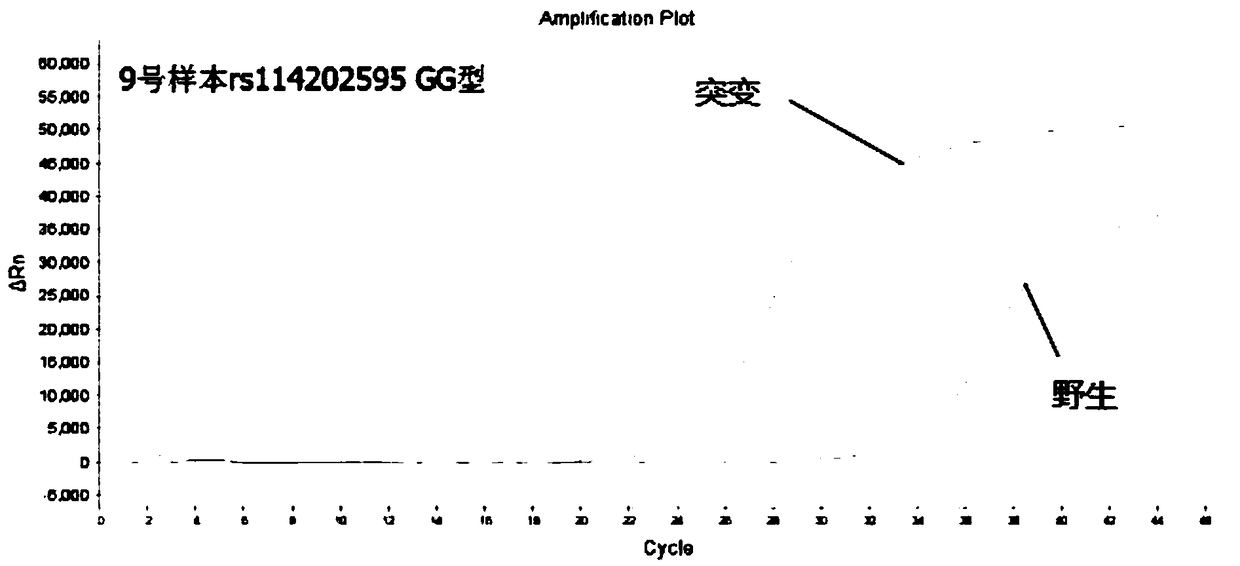
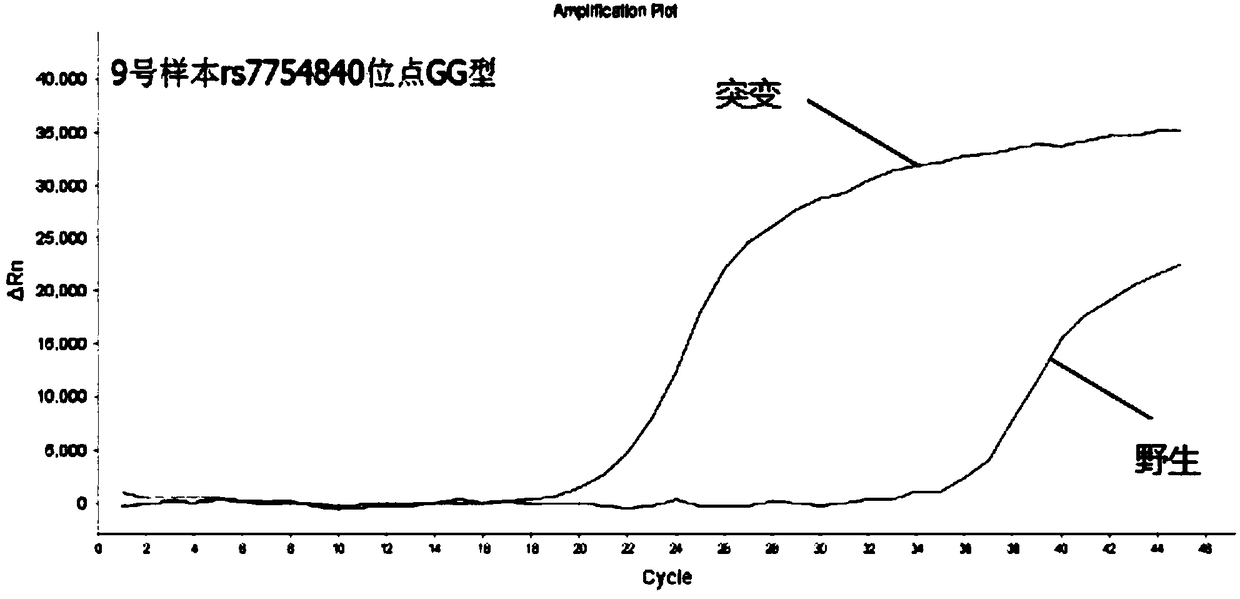
[0107] Embodiment 3: Detect 5 polymorphic sites of diabetes with the kit prepared by the present invention
[0108]For the determination of genomic DNA, almost any biological sample that contains genomic DNA (eg, impure red blood cells) can be used. For example, and without limitation, genomic DNA may be conveniently obtained from blood, semen, saliva, tears, urine, feces, sweat, buccal cells, skin or hair. In order to utilize DNA assays, the target nucleic acid must be obtained from a cell or tissue expressing the target sequence.
[0109] The sample to be tested in this embodiment is DNA extracted from 10 cases of oral swabs collected by Wuhan Liangpei Gene Biotechnology Co., Ltd. and quantified as a template for PCR detection. For the main operation steps of DNA extraction, refer to Tiange Company’s buccal swab DNA extraction kit to extract genomic DNA from samples. The purity and concentration of the extracted genomic DNA were calculated by an ultraviolet spectrophotomet...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com