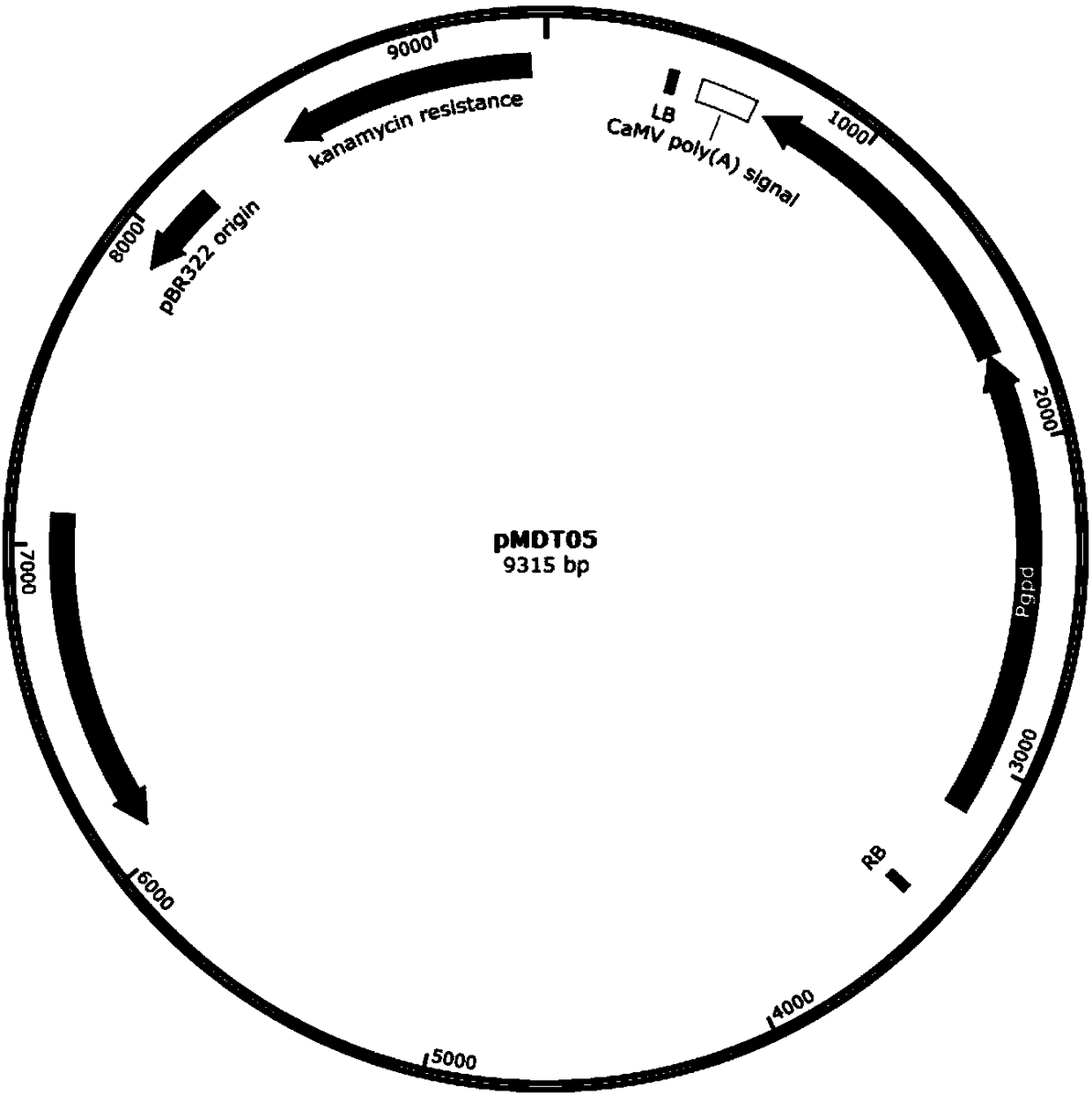
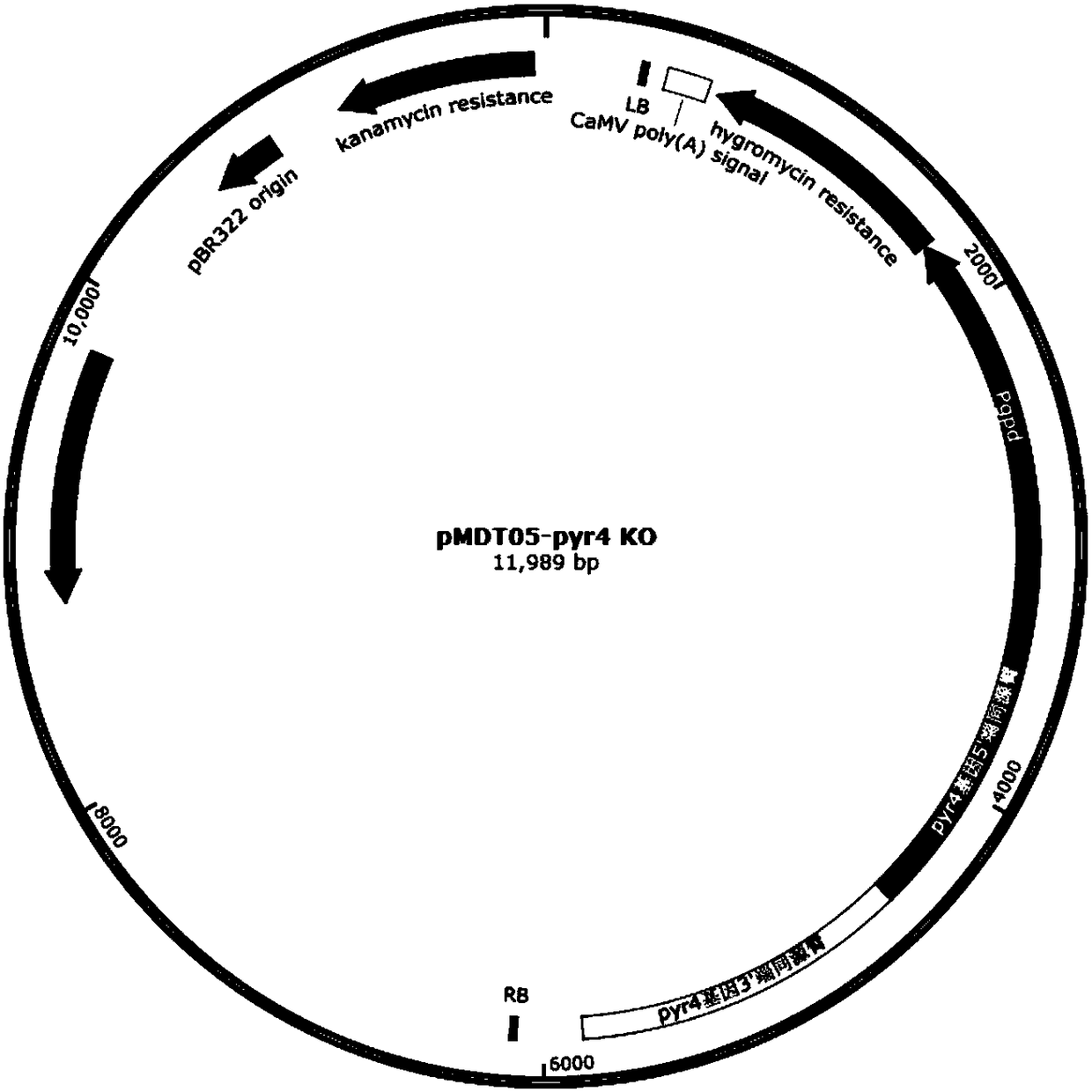
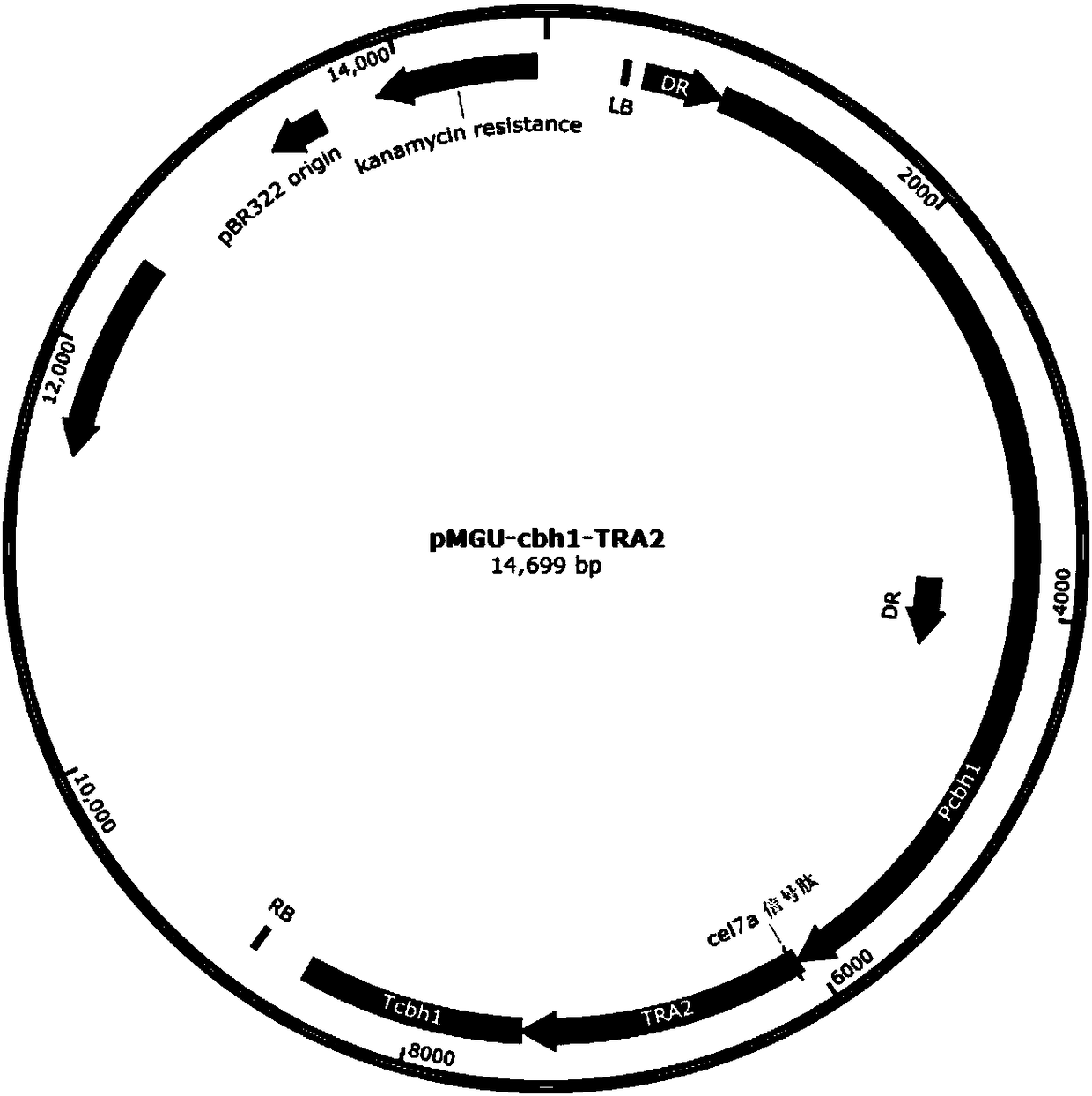
Recombinant oxalate decarboxylase obtained through mycelial fungus host cell expression
A technology of oxalate decarboxylase and host cells, applied in the field of recombinant oxalate decarboxylase, can solve the problem that oxalate decarboxylase cannot be effectively recombinantly expressed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Example 1: Codon optimization and artificial synthesis of oxalate decarboxylase (OXDC) gene
[0080] The present inventors have found through a large number of experimental studies that it can be used in the expression system of filamentous fungi to express oxalate decarboxylase derived from eukaryotes, preferably from tea tree mushroom, poplar mushroom, enoki mushroom, versicolor versicolor, brown rot fungus, aspergillus auricula, bisporus Oxalate decarboxylase from fungi such as mushrooms and jinfu mushrooms.
[0081] The gene encoding oxalate decarboxylase can be derived from Tea Tree Mushroom, the amino acid sequence of the oxalate decarboxylase is the amino acid sequence shown in SEQ ID NO.1, wherein the signal peptide sequence of the oxalate decarboxylase is 1- The 19th amino acid sequence, the mature peptide sequence is the 20-470th amino acid sequence shown in SEQ ID NO.1.
[0082] The OXDC gene derived from A. reesei was optimized according to the codon prefer...
Embodiment 2
[0083]Embodiment 2: Trichoderma reesei Rut-C30 (pyr4 - ) construction of auxotrophic strain
[0084] The filamentous fungal host cells used to express the eukaryotic OXDC in the eukaryotic system are selected from Aspergillus, Coriolus, Mucor, White Rot, Acremonium, Cryptococcus, Fusarium, and Humicola Genus, Myceliophthora, Aureobasidium, Trametes, Pleurotus, Neurospora, Penicillium, Paecilomyces, Phaneroderma, Tobacillus, Cerexe, Thielavia Cells of the genus Coprinus, Chrysosporium, Schizophyllum, Coprinus, Magnaporthe oryzae, Neomystium, Curvularia, Talaromyces, Thermoascomyces, or Trichoderma, or their organisms Sexual or synonymous cells, but not limited thereto.
[0085] The Trichoderma host cell is Trichoderma harzianum, Trichoderma konningen, Trichoderma reesei, Trichoderma longistanum and Trichoderma viride, preferably Trichoderma reesei and Trichoderma viride. Take Trichoderma reesei as an example below to illustrate the present invention.
[0086] 1. Trichoderma...
Embodiment 3
[0109] Example 3: Construction of oxalate decarboxylase random integration recombinant expression vector
[0110] 1. Construction of random integration inducible expression vector pMGU-cbh1-TRA2
[0111] Construction of vector pMGU:
[0112]Using the plasmid vector pMDT05 prepared in Example 2 as a template, use the primers F1 and R1 in Table 3 as the upstream and downstream primers to PCR amplify a vector backbone fragment of about 6.6 kb, use the gel to recover and use the restriction endonuclease DpnI Digest for 3 hours, recover the target fragment, and set aside.
[0113] With reference to the method for Trichoderma reesei genome extraction in Example 2, extract Aspergillus niger CICC2439 genomic DNA, use this genomic DNA as a template, amplify about 2.9kb of Aspergillus niger pyrG with primers pyrG-F and pyrG-R in Table 3 Gene expression frame, gel recovery target fragment, spare. Using Trichoderma reesei genomic DNA as a template, the primers Pcbh-DR-F and Pcbh-DR-R i...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com