Feruloyl esterase and preparing method and application thereof
A technology of ferulic acid esterase and screening method, which is applied in the field of new ferulic acid esterase and its preparation, and the field of preparation of ferulic acid esterase, which can solve the problems that no ferulic acid esterase gene has been found, microorganisms cannot be cultivated, etc. , to achieve good thermal stability and high enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
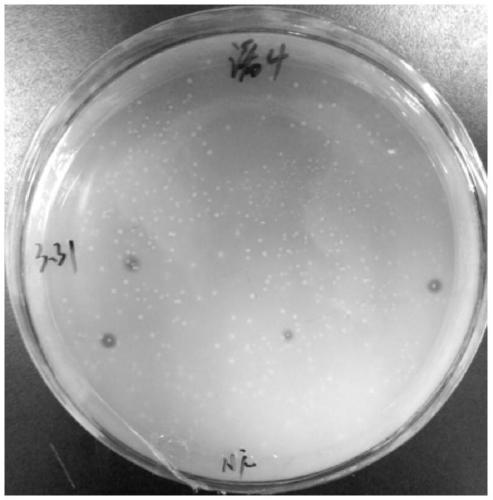
[0053] Example 1: Construction of soil metagenomic library, screening of positive clones and identification of corresponding ferulic acid esterase gene
[0054] Construction of the soil metagenomic library: Weigh 10 g of the soil sample, add CTAB extract, shake and mix at 37°C for 45 minutes. Add appropriate amount of lysozyme and proteinase K to the components to lyse cells and remove proteins. Add 2.5mL of 20% SDS (g / 100mL), bathe in water at 65°C for 2h, then add 3mL of pre-cooled chloroform, mix well and centrifuge to collect the supernatant. Add an equal volume of pre-cooled phenol: chloroform: isoamyl alcohol (25:24:1 volume ratio) solution, invert and mix well and centrifuge, take the upper aqueous phase and add an equal volume of chloroform: isoamyl alcohol (24:1 volume ratio) , centrifuge again, take the water phase and add 0.6 times the volume of pre-cooled isopropanol, ethanol precipitation in a water bath at room temperature for 1 h, and centrifuge to collect the ...
Embodiment 2
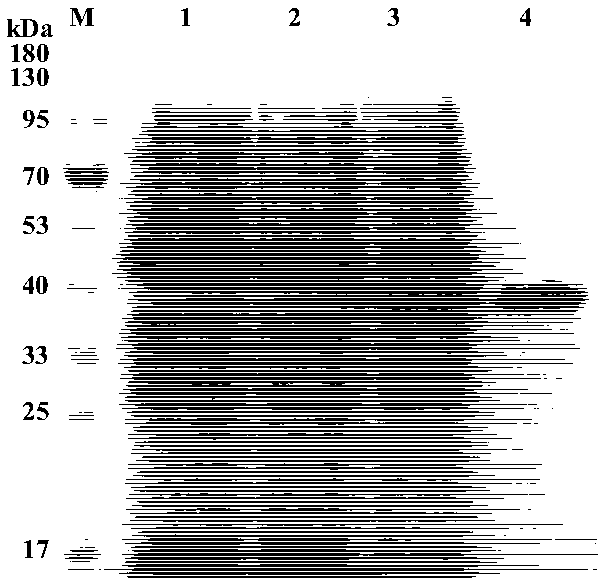
[0058] Embodiment 2: Molecular cloning and expression purification of ferulic acid esterase gene
[0059] Molecular cloning of the ferulic esterase gene: Amplification of the ferulic esterase gene dlfae4fae-F / NcoI:5'-CATG using the following primers CCATGG ATGACGATGGATACG-3' (SEQ ID No. 3) and fae-R / XhoI 5'-CCG CTCGAG TACACTCGCATACACC-3' (SEQ ID No.4) (the restriction sites of NcoI and XhoI are underlined). PCR reaction system (50 μL): 20 μl of ultrapure water, 25 μL of 2×Taq Master Mix, 2 μL of upstream and downstream primers (10 μM), 1 μL of DNA template (the positive clone plasmid DNA prepared in Example 1). PCR reaction conditions: 95°C for 5min; 35 cycles of 95°C for 30s, 56°C for 30s, 72°C for 1min; 72°C for 10min. The PCR product is electrophoresed and recovered by tapping the rubber to obtain a purified ferulic acid esterase gene PCR product. The PCR product was subjected to double enzyme digestion, and the digestion time was 3h. The enzyme digestion system (100 ...
Embodiment 3
[0061] Embodiment 3: Enzymatic property characterization of ferulic acid esterase DLFae4
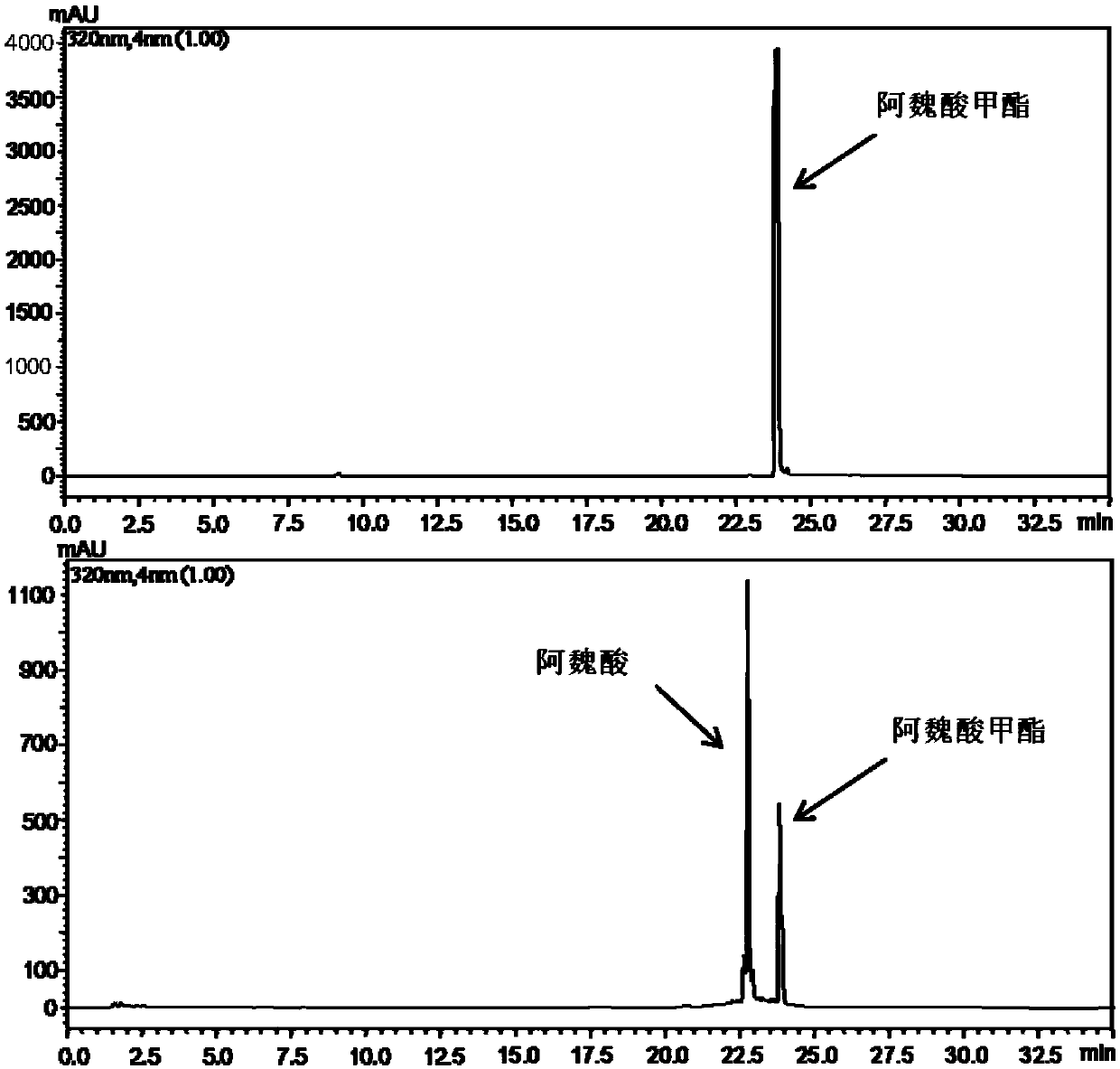
[0062] Determination of enzyme activity: Take 1 mL of Gly-NaOH buffer solution with pH 8.6, add 5 μmol of methyl ferulate solution, add 3 μL of recombinant enzyme, react at 50 ° C, and use HPLC to measure the degradation of the substrate at 320 nm.
[0063] Definition of enzyme activity: under the reaction conditions of 50°C and pH 8.6, the amount of enzyme required to degrade methyl ferulate to generate 1 μmol ferulic acid per minute is defined as 1 enzyme activity unit (U).
[0064] Optimum pH and pH stability analysis: at 37°C, measure the enzyme activity at different pH (3.0-11.0), and determine the optimum pH of the enzyme according to the size of the enzyme activity ( Figure 4 a). Add 3 μL of recombinant enzymes to buffers with different pHs, incubate at 4°C for 3 h, measure the remaining enzyme activity, and determine the stability of the enzyme ( Figure 4 b).
[0065] Optimu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com