Paenibacillus chitin enzyme and application thereof
A technology of Paenibacillus and chitinase, applied in the direction of enzymes, enzymes, hydrolytic enzymes, etc., can solve the problems of insufficient specificity and limited use, and achieve the effects of reduced crystallinity, improved hydrolysis efficiency, and huge application potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
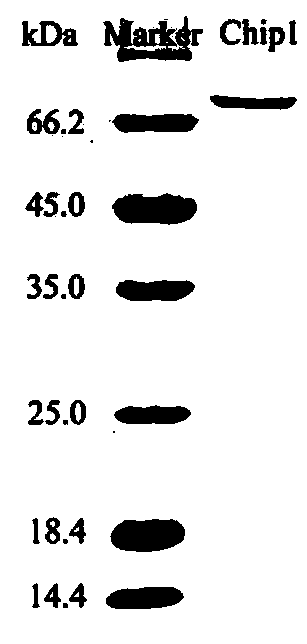
[0035] Example 1 Paenibacillus chitinase Pp Cloning, expression, isolation and purification of Chi2
[0036] (1) Pp Cloning of Chi2
[0037] According to the results of genome sequencing, select the gene containing the signal peptide sequence Pp The gene sequence of Chi2 (Genebank No.: KX431049.1) was used as the research object, and the forward primer Chip1_F: 5'-GGAATTC was designed based on the prediction results of the signal peptide sequence CATATG GCGGCCTGGACGCCCGGC-3', reverse primer Chip1_R: 5'-GACCG CTCGAG GCCGAAGTAGCCCTTCATCGCGT-3', with Paenibacillus pasadenensis The genome of CS0611 was used as a template and amplified Pp Chi2.
[0038] The PCR reaction system is: 10 μL 5X Q5 reaction buffer, 1 μL 10 mM dNTPs, 2.5 μL 10 μM Chip2_F and Chip2_R, 1 μL genomic DNA, 0.5 μL Q5 high-fidelity DNA polymerase, 10 μL 5X Q5 high GCenhancer, 23 μL ddH2O.
[0039] The PCR reaction conditions were: pre-denaturation at 98°C for 30s, denaturation at 98°C for 10s, anneali...
Embodiment 2
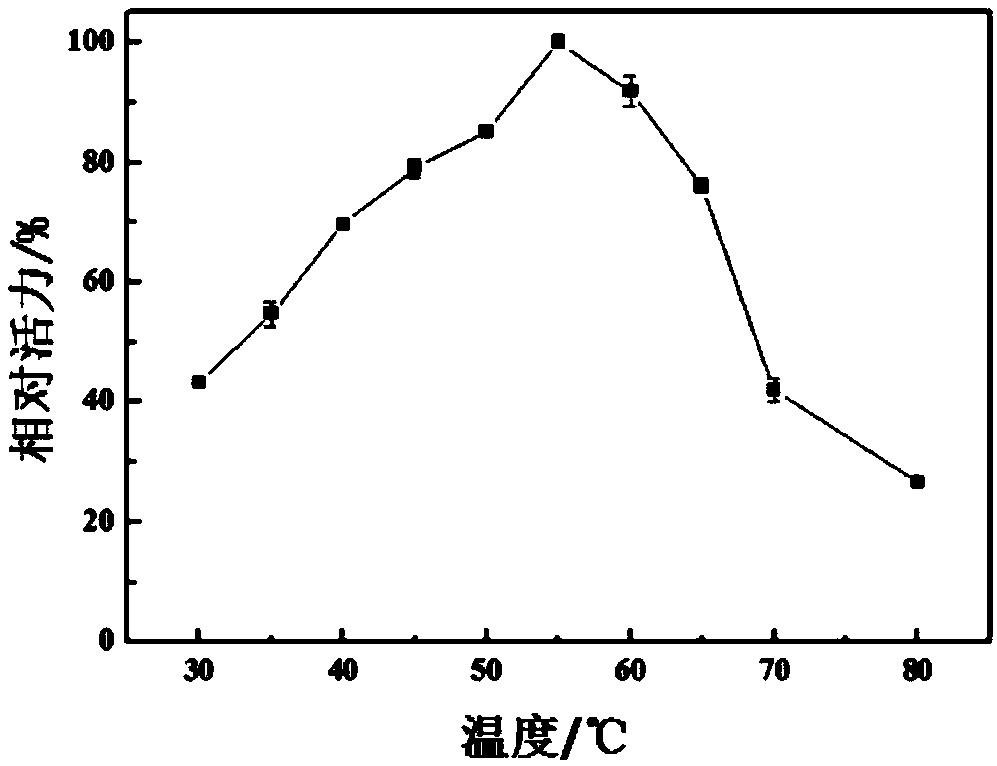
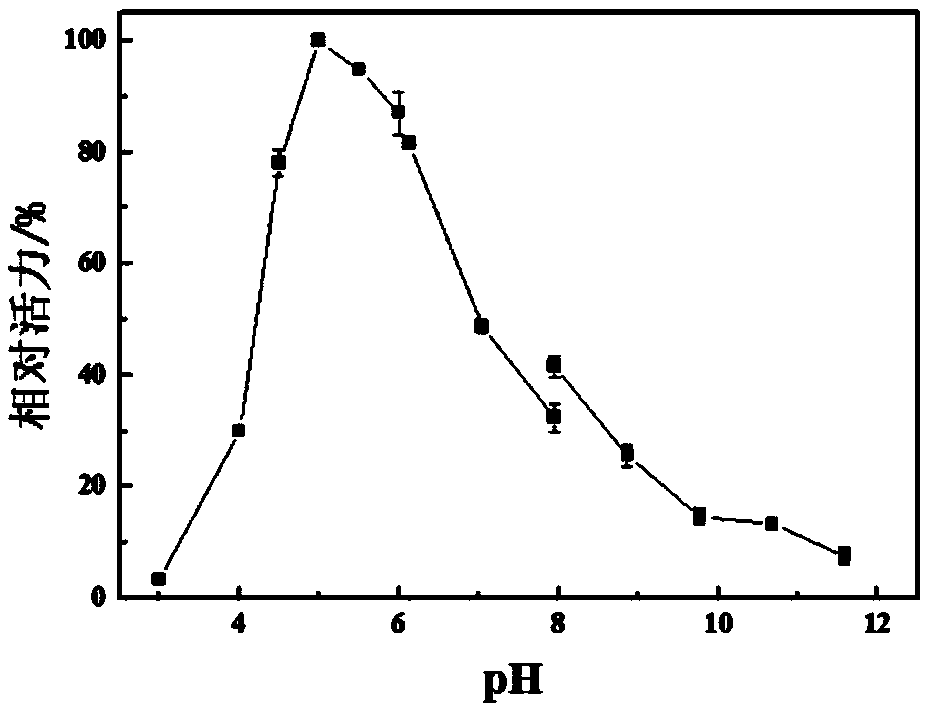
[0047] Example 2 temperature and pH pair Pp Effect of Chi2 Enzyme Activity
[0048] Determination of chitinase activity: the determination of chitinase activity adopts the method of DNS to determine the reducing sugar in the reaction product, and the substrate is 2 wt% colloidal chitin solution. Add 2mL of DNS reagent to 1mL of the reaction product, place in a boiling water bath for 5min, cool down, add water to 10mL, and measure its absorbance at 540nm. The standard curve was drawn with N-acetylglucosamine as the standard substrate, and the unit of enzyme activity was defined as one unit of 1 μM N-acetylglucosamine produced per minute.
[0049] The influence of temperature: 0.5mL respectively Pp Chi2 solution (0.1 mg / mL, 5mM pH7.0 PBS) and 0.5 mL 2wt% colloidal chitin solution (pH 5.0 0.2 M citric acid buffer) were added to a 10 mL centrifuge tube, and then the reaction system was placed at different temperatures (30 , 35, 40, 45, 50, 55, 60, 65, 70, 80 ℃), magnetic stirri...
Embodiment 3
[0051] Embodiment 3 Different proportions Pp Chi1 and Pp Crab shell powder after Chi2 synergistic hydrolysis pretreatment
[0052] Pp Chi1 is and Pp Another chitinase derived from the same target strain with different Chi2 properties, this example examines different mass ratios Pp Chi1 and Pp Chi2 synergistic hydrolysis of crab shell powder pretreated with ionic liquids.
[0053] Pp Chi1 and Pp Chi2 was prepared by the method in Example 1, Pp The concentration of Chi1 is 3.3mg / mL, Pp The concentration of Chi2 was 2.9 mg / mL. The substrate used is crab shell powder (treated-CSP) pretreated by ionic liquid (1-butyl-3-methylimidazolium acetate). Add the powder into 2mL 1-butyl-3-methylimidazole acetate, incubate at 100°C for 1 hour, add water to precipitate chitin, and separate the precipitate by suction filtration. The resulting precipitate was added to 20 mL of deionized water, ultrasonicated for 5 min, and the precipitate was separated by suction filtration. The abov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com