Kit for rapid test of pseudomonas aeruginosa
A Pseudomonas aeruginosa and kit technology, applied in the field of molecular biology, can solve the problems of inability to meet the needs of rapid and simple instant detection, high requirements for experimental conditions, and high maintenance costs, and achieve real-time detection of Pseudomonas aeruginosa. , The amplification compatibility is strong, and the effect of eliminating pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Primer design
[0080] According to the Pseudomonas aeruginosa toxA gene sequence published by NCBI (Reference Sequence: NC_002516.2), based on the principle of enzyme strand displacement activity, a specific primer pair for rapid detection of Pseudomonas aeruginosa, and the nucleoside of the primer pair were designed The acid sequence is as follows:
[0081] Ft: 5'- gcatcgtcttcggcggggt GGAGCGCGGCTATGTGT-3'; SEQ ID NO.1;
[0082] Bt: 5'- gcatcgtcttcggcggggt TCGGGTTCCTGGTCCTG-3'; SEQ ID NO. 2.
[0083] In this embodiment, Chelex-100, betaine, Sigma company; Manganese chloride, magnesium sulfate, potassium chloride, sodium hydroxide, EDTA, ammonium sulfate, Sinopharm Chemical Reagent Co., Ltd.; Tris-HCl, Shanghai Shengke Biotechnology Co., Ltd.; TritonX-100, Beijing Meilaibo Medical Technology Co., Ltd.; dNTP, Pharmacia Company; NP-40, Fluka Company; 2×TapMIX kit, Tiangen Biochemical Technology (Beijing) Co., Ltd.; Agarose, Amresco Company.
[0084] The reagent in th...
Embodiment 2
[0090] Accelerate the introduction of primers
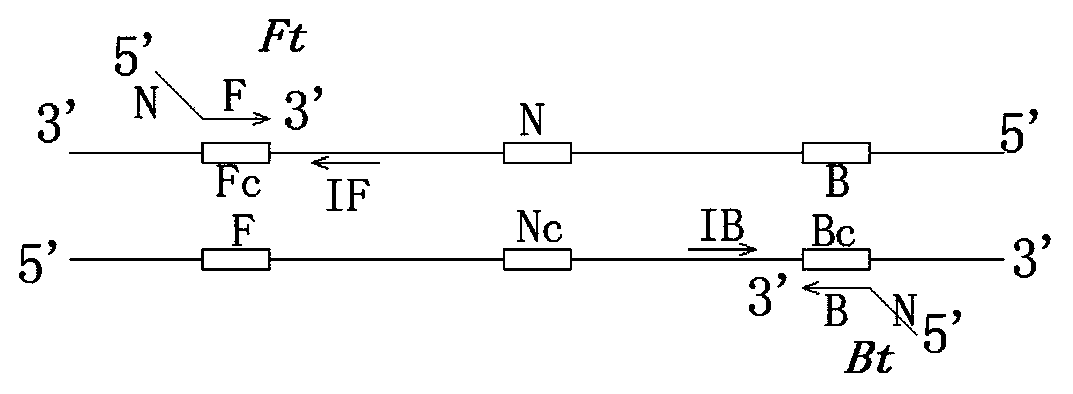
[0091] If only two primers are used, the peaking time is generally after 50 minutes, which cannot meet the requirements of rapid and sensitive on-site detection. The introduction of accelerated primers includes IF and IB, respectively, between F and N, B and N, starting from the 5' end The direction to the 3' end is opposite to Ft and Bt respectively, and the schematic diagram of the positions of IF and IB on the target sequence is shown in figure 2 .
[0092] In this example, the accelerating primer is IF: 5'-AAGGTGCCGTGGTAGCC-3'; SEQ ID NO.3;
[0093] IB: 5'-TCTATATCGCCGGCGATCC-3'; SEQ ID NO.4.
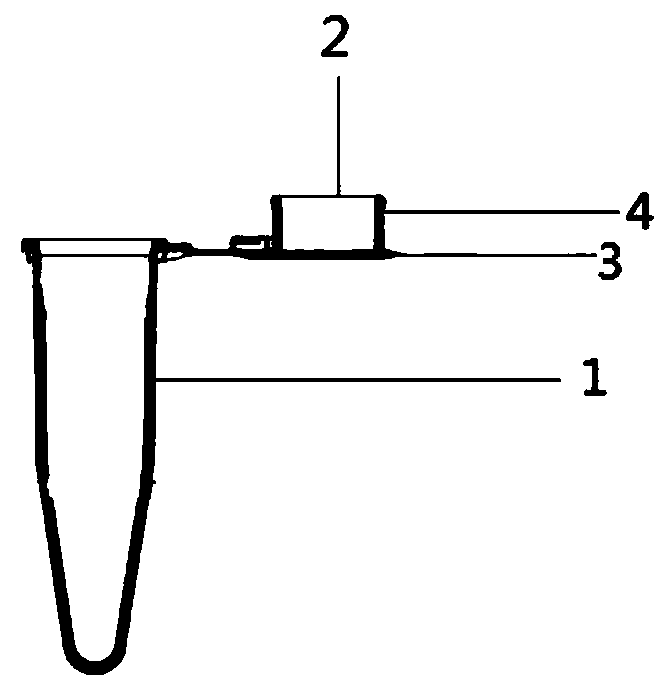
[0094] The reagents in the kit of the present invention are stored using a one-pack diagnostic reagent bottle, wherein the reagent bottle is provided with a detection tube 1, a detection tube cover 3, a placement groove 4 and a coating 2; wherein the detection tube 1 is open at one end , a hollow reaction tube with the other end cl...
Embodiment 3
[0098] Betaine concentration is verified on the basis of Example 2.
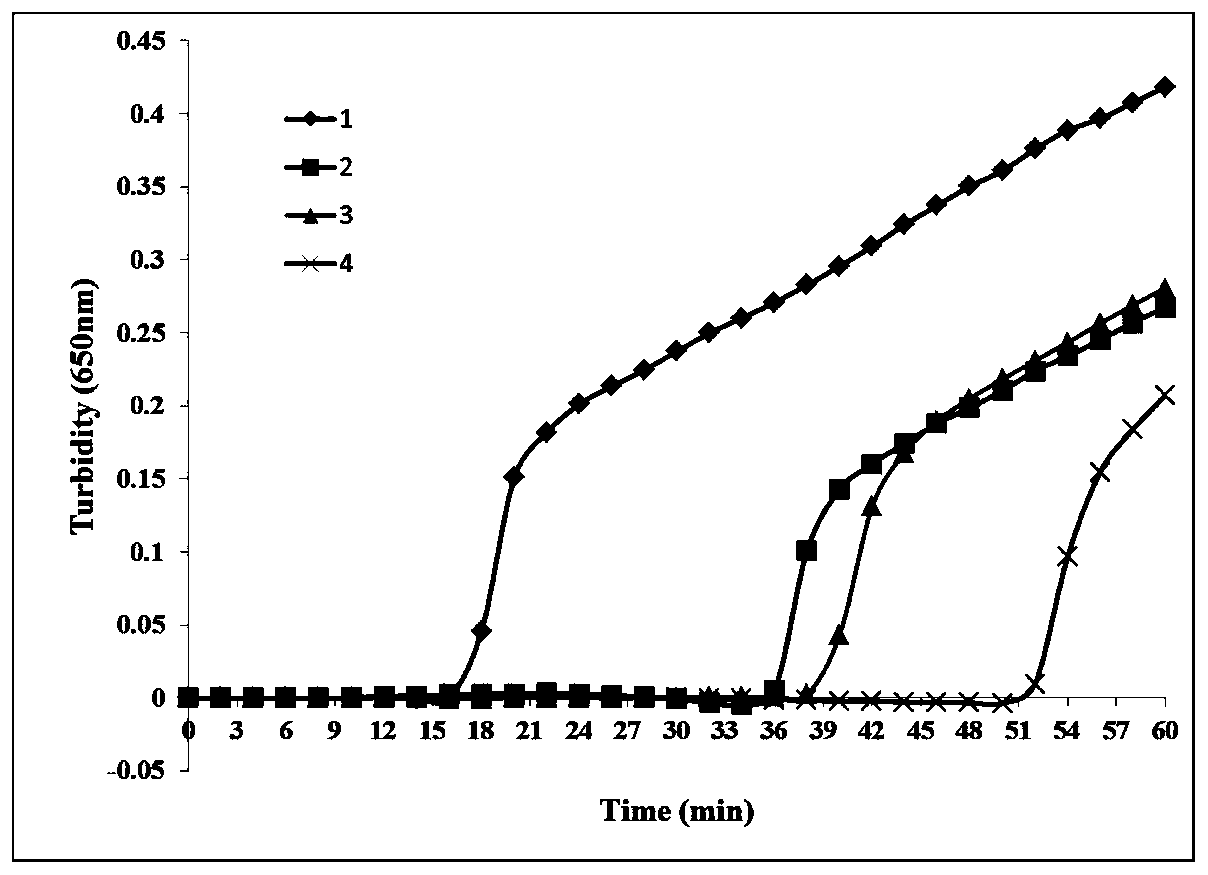
[0099] Set seven betaine concentrations (working concentrations) from 0.5M to 1.1M with a spacing of 0.1 to investigate the influence of different betaine concentrations on the reaction Ct value.
[0100] Detected by turbidity method, the results are as follows Figure 4 As shown, No. 3 curve (0.7M) and No. 4 curve (0.8M) have the fastest reaction peak rate, but the peak concentration of betaine at 0.8M is slightly higher than 0.7M, and finally 0.8M is used as its optimum concentration.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com