Moringa seed protease capable of promoting goat milk solidification and polypeptide prepared from moringa seed protease and application
A technology of Moringa oleifera seeds and protease, applied in the field of dairy science, can solve the problems of unpredictable polypeptide functional activity, inability to guarantee added value, few reports on the research of protease curd function, etc., to achieve the effect of enhancing functional activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
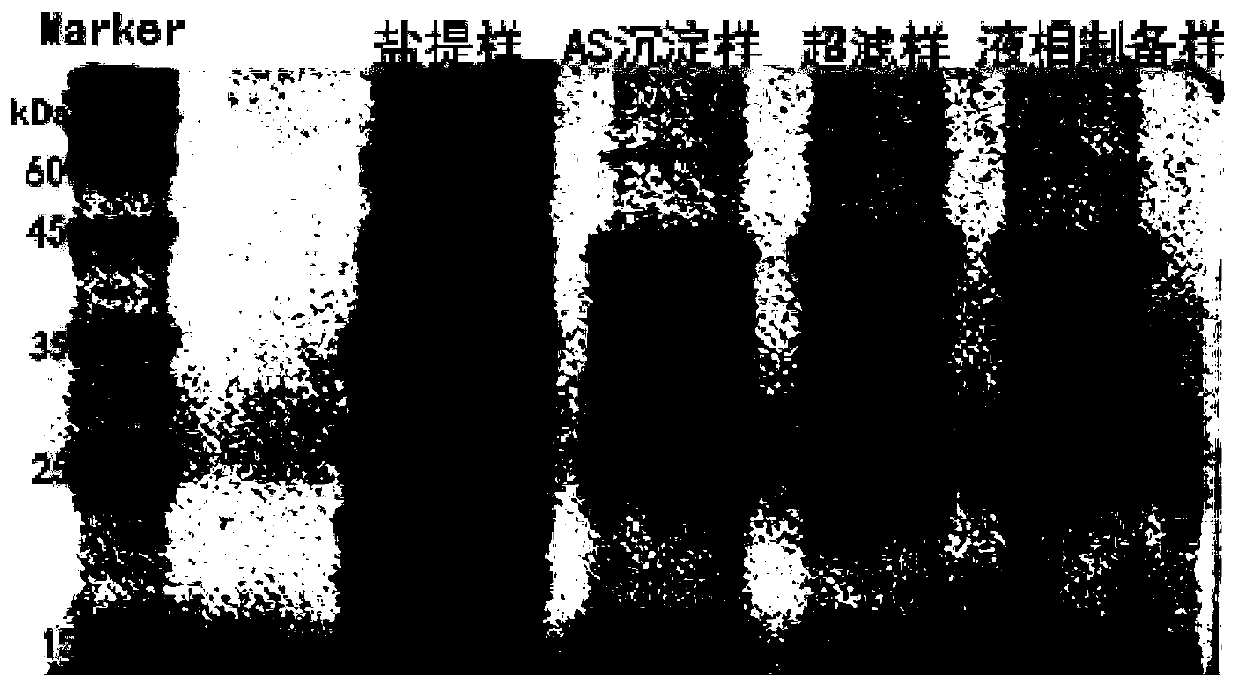
[0032] The extraction of embodiment 1 Moringa seed protease
[0033] 1) Get 100 grams of Moringa seeds (Indian species PKm2, tree age 3 years), the Moringa seeds moisture content is 10.5%, the Moringa seeds are obtained through mechanical pulverization to obtain the Moringa seeds powder, then extract the Moringa seeds by supercritical carbon dioxide The powder is degreased. The conditions for degreasing treatment of Moringa oleifera seed powder by supercritical carbon dioxide extraction are: under the conditions of extraction pressure 30MPa, extraction temperature 40°C, and separation temperature 40°C, 26.7% of the fat in Moringa oleifera seeds is removed, and the degreasing rate reaches 72.8%.
[0034] 2) Extraction of Moringa seed crude protein: use the salt method to process the defatted Moringa seed powder to obtain the crude protein extract of Moringa seed; the optimal extraction process parameters are solid-liquid ratio 1:10, extraction temperature 30 ℃, pH=7.15, salt c...
Embodiment 2
[0047] The identification of embodiment 2 Moringa seed protease
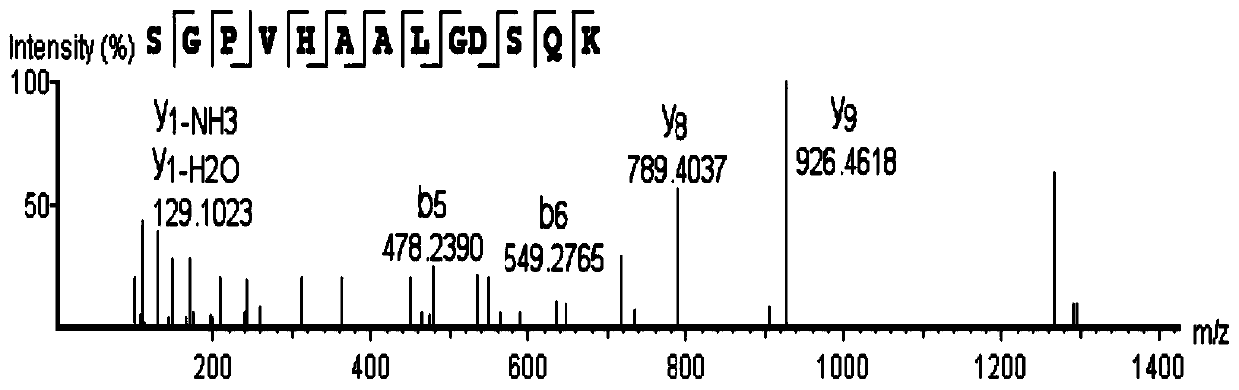
[0048] 1) Reductive alkylation treatment (10mM TCEP[Tris(2-carboxyethyl)phosphine] at 37°C for 60 minutes, 40mM iodoacetamide at room temperature for 40 minutes) was performed on the purified Moringa seed protease, followed by enzymatic After treatment (trypsin, 18h), peptides were extracted for LC-MS / MS. Use the PEAKS Studio 8.5 software to retrieve data from the Moringa oleifera transcriptome database to identify the Moringa oleifera seed protease, and the identification results are as follows figure 2 As shown in the figure, the ion selection device automatically selects peptide ions with higher intensity to simulate fragmentation (secondary fragment ion spectrum matching)-mass spectrum prediction scoring evaluation. The peptide score will be calculated based on the size of the fragmented ions in the computer simulation and the matching degree in the actual secondary spectrum, and the one with the highest m...
Embodiment 3
[0053] Example 3 The cleavage site of κ-casein digested by Moringa oleifera seed protease and the polypeptide produced by enzymatic digestion
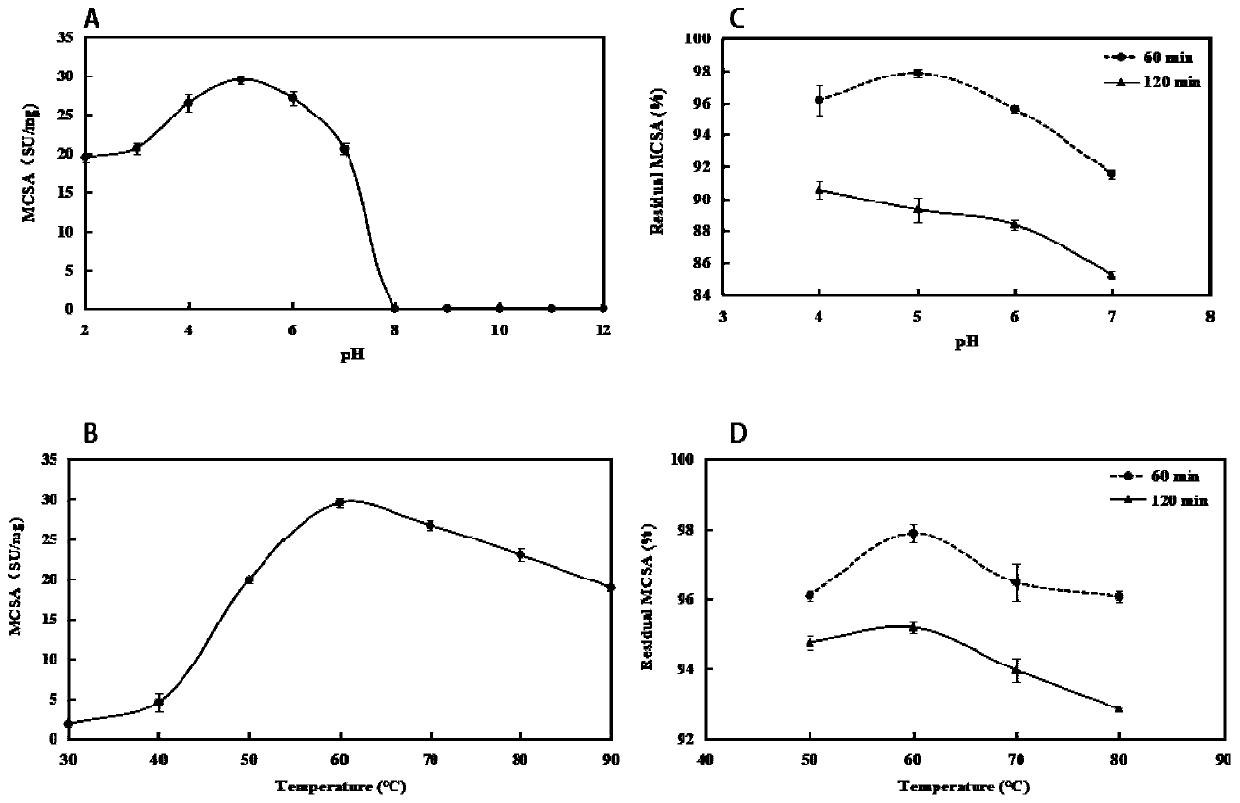
[0054] 1) Use the purified Moringa oleifera protease and κ-casein (κ-CN) at a ratio of 1:10 to react in a water bath at 60°C for 60 minutes, and then centrifuge the obtained protein solution through a Millipore ultrafiltration centrifuge tube (molecular weight 3k Da) at 4°C After desalting and removing impurities, the protein gel strips in which Moringa oleifera seed protease interacted with κ-casein for 60 minutes were obtained by SDS-PAGE, and the gel was cut for matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF / TOF), and the molecular weight, sequence and restriction sites.
[0055] The amino acid sequence of κ-casein is as follows. The amino acid sequence information of κ-casein comes from NCBI database, and the species source is bovine:
[0056]
[0057]
[0058] The digestion site is G...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com