PET degradation biocatalyst and its application
A catalyst and enzyme-degrading technology, applied in biochemical cleaning devices, enzyme production/bioreactors, biochemical equipment and methods, etc., can solve the problems of difficult separation and recycling of blended fibers, secondary pollution of the environment, expensive catalysts, etc., to achieve Significant economy and high efficiency, low reaction temperature, and reduced process cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1: Plasmid expresses PET degrading enzyme
[0037] The genes encoding PET degrading enzymes such as LCC (SEQ NO.1), Tcur (serial number ACY96861.1), PETase (serial number GAP38373.1), Tcut (serial number WP_011291330) were selected for expression in Clostridium thermocellum DSM1313, Construction of PET degradation biocatalyst. The specific steps are:
[0038] 1) Connect the signal peptide sequence (SEQ NO. 2) to the 5' end of the above gene by seamless cloning.
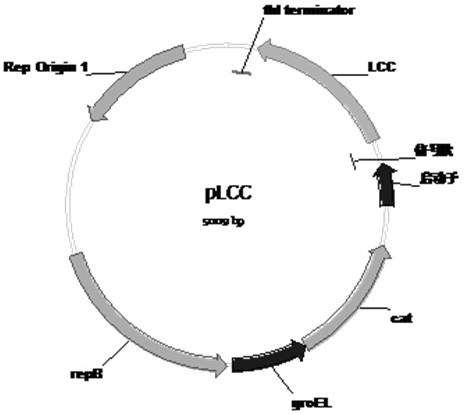
[0039] 2) Connect the sequence obtained in step 1) to the 3' end of the promoter I of the pHK plasmid using the seamless cloning method. Wherein pHK is a shuttle plasmid with replicons of Escherichia coli and Clostridium at the same time, with chloramphenicol and thiamphenicol resistance genes; the promoter I sequence is SEQ NO. 3, and the obtained PET degrading enzyme expression plasmid is shown in figure 1 . pLCC is a plasmid expressing LCC; pTcur is a plasmid expressing Tcur; pPETase is a pla...
Embodiment 2
[0042] Example 2: Genomic expression of PET degrading enzymes
[0043] (1) Using the method of seamless cloning, sequentially connect the nucleic acid sequences of promoter I, signal peptide, LCC and MHETase in SEQ NO.3, SEQ NO.2, SEQ NO.1 and SEQ NO.9 to form a fusion expression box, wherein the stop codon of LCC is removed.
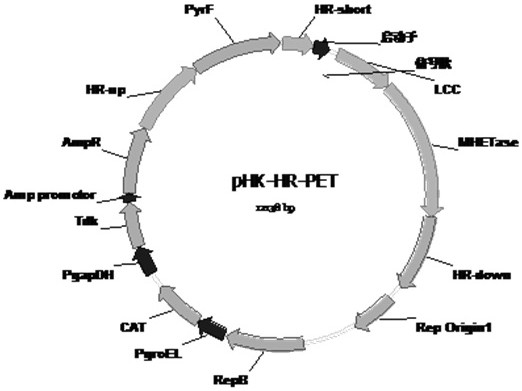
[0044] (2) Select the 16SrRNA gene sequence as the target site, and clone the above fusion expression cassette as the target gene into the homologous recombination plasmid pHK-HR (Reference 1) to obtain the genomic recombination plasmid pHK-HR-PET ( figure 2 ). Using the scarless knockout and screening method (document 1), it was integrated into the genome of Clostridium thermocellum, and strains expressing LCC and MHETase were obtained by genome fusion.
[0045] (3) After the mutant strain is obtained, the enzyme activity of the extracellular protein with pNPB as the substrate is determined to confirm whether the PET degrading enzyme is successfully...
Embodiment 3
[0046] Embodiment 3: express PET degradation body:
[0047] (a) The plasmid pLa-Mf co-transcriptionally expressing LCC and MHETase was constructed, and the two enzymes were respectively fused with assembly modules from Clostridium acetobutylicum and Clostridium flavum. Specifically: the expression assembly module DocCa (SEQ NO.4) was fused at the 3' end of the LCC coding gene of the above pLCC plasmid to obtain plasmid pLa; the RBS sequence (AGGAGG) and the coding gene of MHETase ( SEQ NO.9) and the gene of the assembly module DocCf (SEQ NO.5) to obtain the plasmid pLDa-MDc.
[0048] (b) Cloning the co-transcriptional expression cassette in pLDa-MDf as the target gene into the homologous recombination plasmid pHK-HR (Reference 1), selecting the 16S rRNA gene sequence as the target site, and using the scarless knockout and screening method (Reference 1) 1), integrated into the genome of Clostridium thermocellum, to obtain strains co-transcribing and expressing LCC and MHETase ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com