Application of gene SNG1 deletion in improving vanillic aldehyde resistance of saccharomyces cerevisiae
A technology of Saccharomyces cerevisiae and gene deletion, applied in the application, genetic engineering, plant genetic improvement and other directions, to achieve the effect of shortening the fermentation cycle, improving resistance and saving production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1: Construction of sng1Δ mutant
[0046] (1) Extraction of Saccharomyces cerevisiae BY4741 genome
[0047] Inoculate Saccharomyces cerevisiae BY4741 in 5mL YEPD medium, cultivate overnight at 30°C, 200rp, collect the bacteria by centrifugation, wash once with 1mL of sterile water, add 200μL of lysis buffer to suspend the cells, and transfer to a cell containing 0.4g of glass beads. Add 200 μL of phenol / chloroform / isoamyl alcohol (25:24:1) to the screw-top broken cell tube, vortex for 3 minutes, add 200 μL of TE solution to mix, centrifuge at 13000 r / min for 10 minutes at 4 °C, transfer to Clear to a new Eppendorf tube, add 1ml of -20°C absolute ethanol to precipitate DNA, then centrifuge at 13000r / min for 10min at 4°C, and discard the supernatant. After vacuum drying, add 400 μL of TE solution to dissolve DNA, then add 10 μL of 10 mg mL -1 RNase A, after incubation at 37°C for 15min, add 10μL of 4mol L -1 After mixing the ammonium acetate and 1mL of absolute ...
Embodiment 2
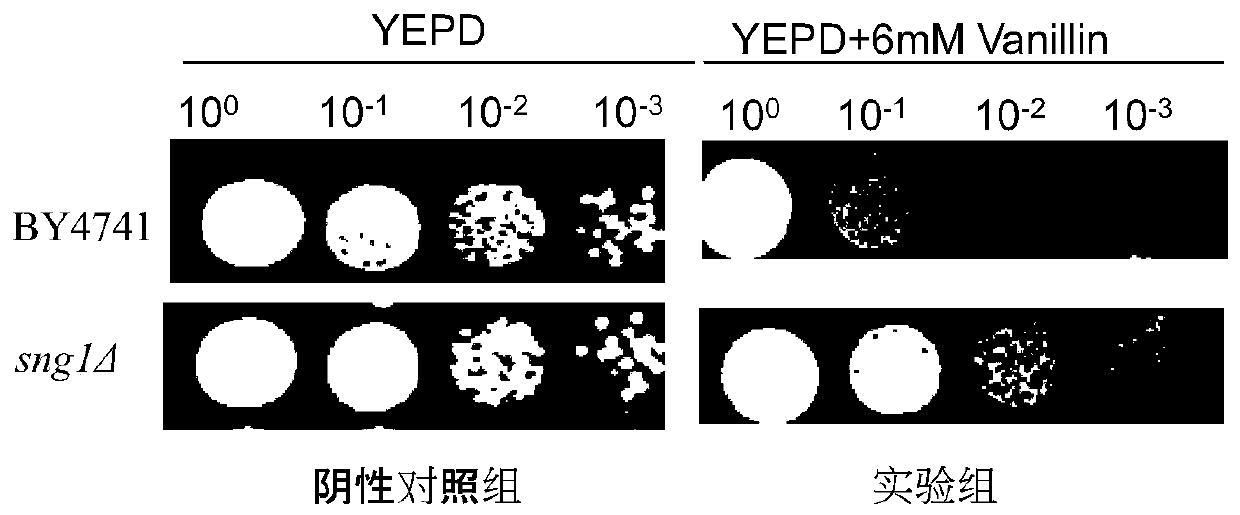
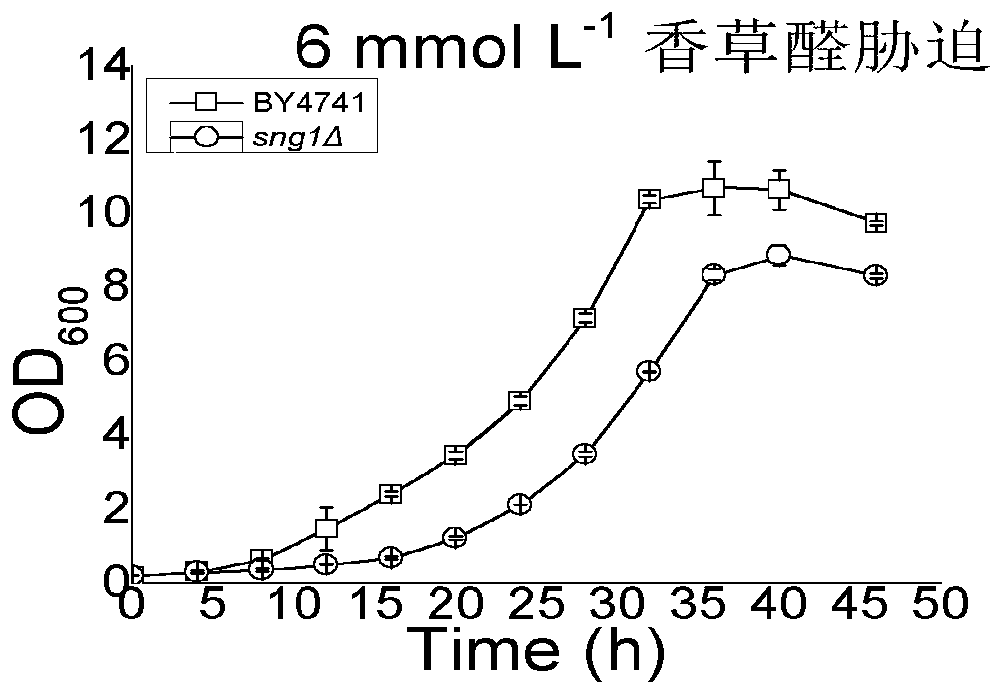
[0089] Example 2: Application of sng1Δ mutant in improving vanillin resistance (vanillin tolerance)
[0090] (1) Vanillin tolerance experiment of sng1Δ mutant
[0091] The obtained sng1Δ mutant was mixed with 6mmol L -1 Vanillin gradient growth experiments were performed on YEPD plates. The specific operation is as follows:
[0092] Inoculate the sng1Δ mutant in 1-2mL of YEPD medium, culture overnight at 30°C until the logarithmic growth phase, and the culture product was measured at the initial OD 600 Transfer 0.2 to 5mL fresh culture medium, collect the cells by centrifugation after cultivating overnight, wash the suspended cells with 1mL sterile water for 3 times, centrifuge at 8000r / min, suspend the cells with sterile water again, put them in Placed at 30°C for 9h to prepare resting cells. During the preparation of the culture medium, pour it on the plate and let it dry naturally. Adjust the concentration of bacterial suspension to OD 600 1.0, followed by 10-fold ser...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com