A kind of chondroitin sulfate lyase and its encoding gene and application
A technology of chondroitin sulfate and coding genes, which is applied in the field of enzyme engineering to achieve the effects of high degradation activity, easy heterologous expression and purification, and stable physical and chemical properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
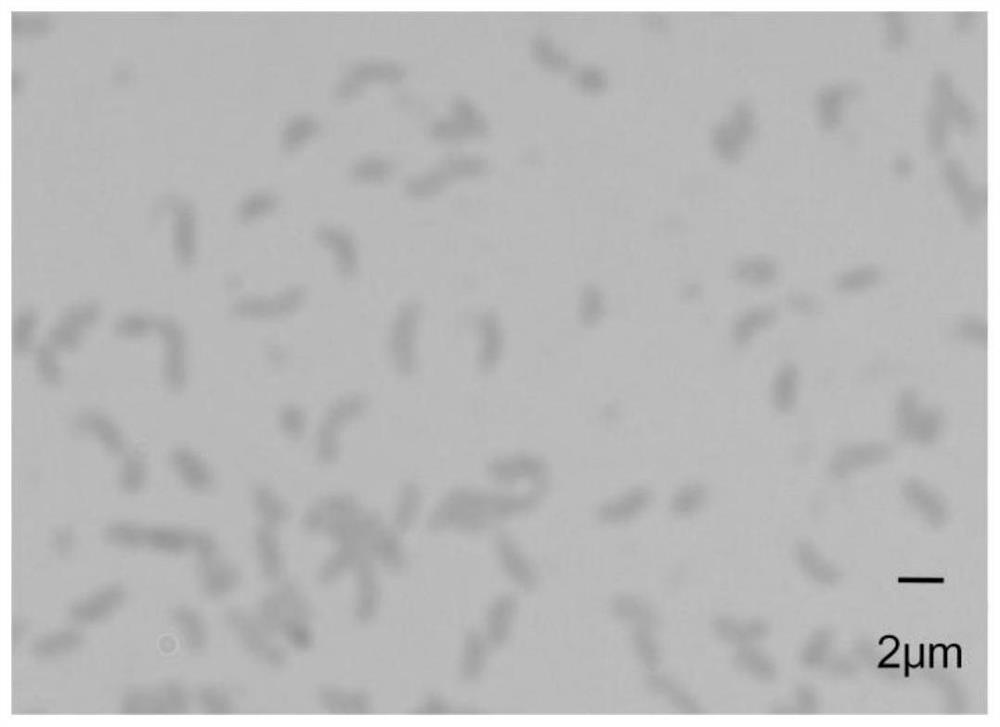
[0042] Example 1. Obtaining of Photobacterium sp. QA16
[0043] Take sea mud leachate, add 1 mL of supernatant to 9 mL of sterile water, and dilute to 10 -1 , 10 -2 , 10 -3 , 10 -4 , 10 -5 , 10 -6 Then, by conventional dilution method or streaking method, it was inoculated on the sole carbon source solid medium, incubated at 30°C for 1 day, the colonies were counted, and then the colonies with obvious differences in colony morphology were selected, and the streaking was repeated. It was inoculated on the corresponding complete nutrient agar plate until a single colony was obtained after purification, and then transferred to the corresponding agar slant for later use.
[0044] The cultured strains were respectively inoculated onto the only carbon source liquid medium for cultivation. Incubate at 200rpm and 30°C for 72h, observe the turbidity of the bacterial liquid, and take the culture supernatant for carbazole reaction to detect the consumption of carbon source. The en...
Embodiment 2
[0049] Example 2. Extraction of genomic DNA of Photobacterium sp. QA16
[0050] Photobacterium sp. QA16 was inoculated into the liquid medium and cultured to OD with shaking at 30°C and 200rpm. 600 About 0.8; take 40 mL of cultured bacterial solution, centrifuge at 12,000 rpm for 25 min, collect the bacterial pellet, wash with 20 mL of lysozyme buffer (10 mM Tris-HCl pH 8.0), and centrifuge at 12,000 rpm for 25 min to collect bacteria. body precipitation;
[0051] In the above-mentioned cell precipitation, add 12.0 mL of lysozyme buffer (10 mM Tris-HCl pH 8.0) to each tube to obtain about 14.0 mL of bacterial liquid, respectively add 560 μL of lysozyme with a concentration of 20 mg / mL, and its final concentration is about 800 μg / mL; after ice bath for 1.0h, incubate at 37°C for 2h until the solution is viscous; add 0.82mL of 10wt% SDS (sodium dodecyl sulfonate), 60μL of 100mg / mL proteinase K solution, water bath at 52°C for 1.0h; Add 15mL of Tris-equilibrated phenol / chlorof...
Embodiment 3
[0052] Example 3. Genome scanning and sequence analysis of Photobacterium sp. QA16 strain
[0053] The large molecular weight genomic DNA obtained in Example 2 was sequenced (Beijing Yuanyi). The sequencing results were analyzed with software on NCBI (National Center for Biotechnology Information, http: / / www.ncb1.nlm.nih.gov / ). The NCBI analysis software used is Open Reading Frame Finder (ORF Finder, http: / / www.ncb1.nlm.nih.gov / gorf / gorf.html) and Basic Local Alignment SearchTool (BLAST, http: / / blast.ncb1 .nlm.nih.gov / Blast.cgi).
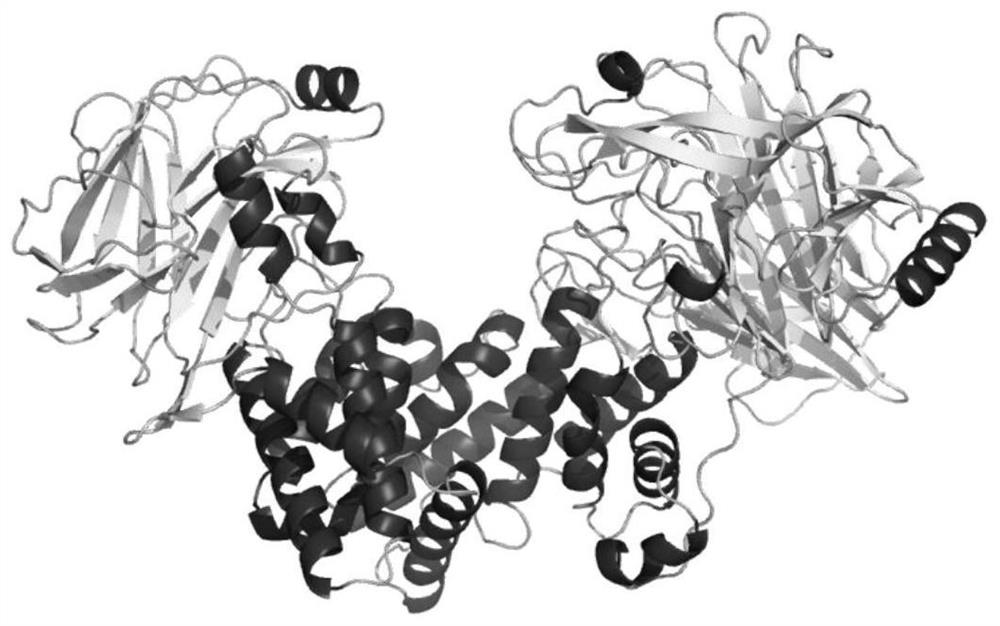
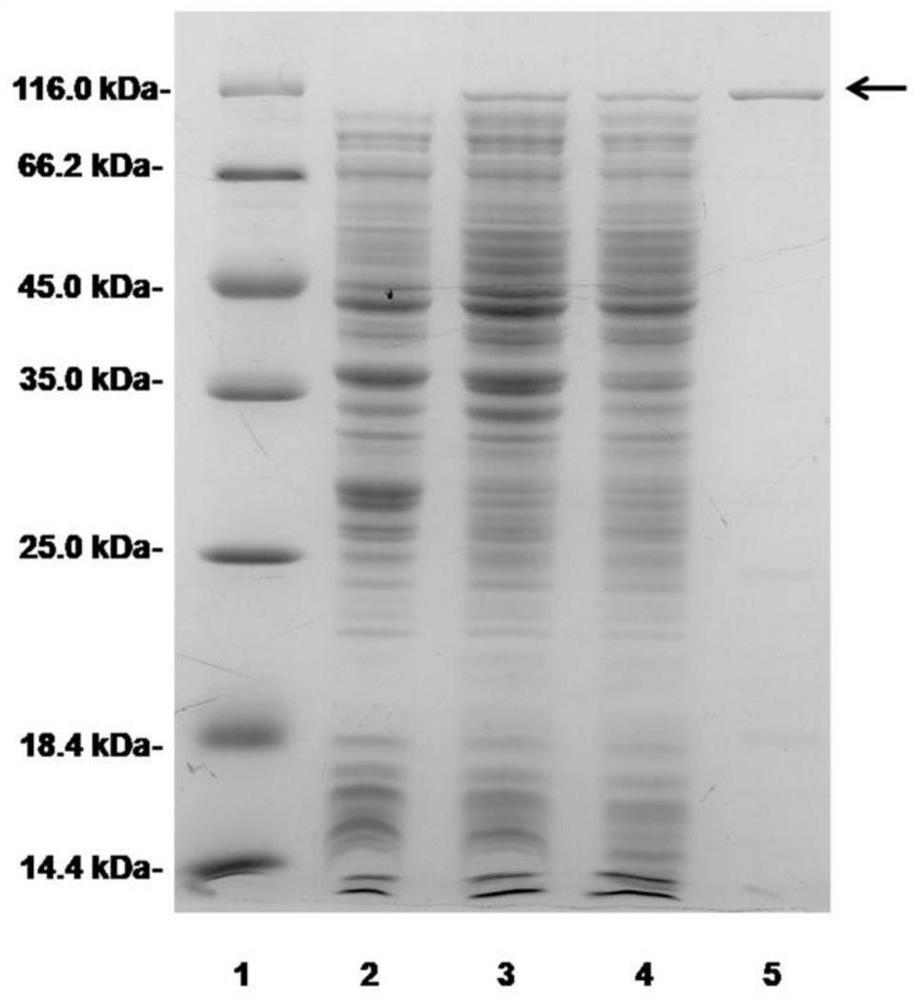
[0054] The NCBI analysis results showed that the chondroitin sulfate lyase gene encsase carried on the genome of Photobacterium sp. QA16 strain has a length of 2943 bp in the gene coding region, and its nucleotide sequence is shown in SEQ ID NO.1. The chondroitin sulfate lyase enCSase encoded by the chondroitin sulfate lyase gene encsase consists of 980 amino acids, the amino acid sequence of which is shown in SEQ ID NO. 2, and the theoretical mol...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com