Method for synthesizing methyl selenocysteine by using bacillus subtilis
A technology of methylselenocysteine and Bacillus subtilis, which is applied in the field of bioengineering, can solve the problems of complex steps, lengthy process, and shortage, and achieve the effects of reducing production costs, convenient use, and enhancing secretion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1: Construction of recombinant Bacillus subtilis / pP43NMK-SMT
[0037] (1) Construction of expression vector pP43NMK-SMT
[0038] Chemically synthesize the gene encoding SMT protein (nucleotide sequence shown in SEQ ID NO.1); add pP43NMK homologous sequence (nucleotide sequence shown in SEQ ID NO.13 and The nucleotide sequence is shown in SEQID NO.14) to obtain the SMT fragment; the pP43NMK was digested and recovered by Kpn I and Sma I to obtain the pP43NMK fragment; the SMT fragment and the pP43NMK fragment were assembled using Gibson Assembly Master Mix to obtain the SMT expression vector pP43NMK -SMT.
[0039] Transform the expression vector pP43NMK-SMT into competent cells to obtain transformation products; spread the transformation products in LB culture to obtain transformants, pick the transformants and inoculate them in LB medium for culture, extract plasmids for enzyme digestion verification and sequencing verification , the verification is correct ...
Embodiment 2
[0042] Embodiment 2: Construction of cysteine transporter expression vector
[0043] (1) Extraction of Escherichia coli genomic DNA: Inoculate Escherichia coli strains into LB medium, culture at 200r / min at 37°C for 12h, collect the bacteria by centrifugation, and extract the genomic DNA of Escherichia coli using the bacterial genomic DNA kit of TIANGEN company .
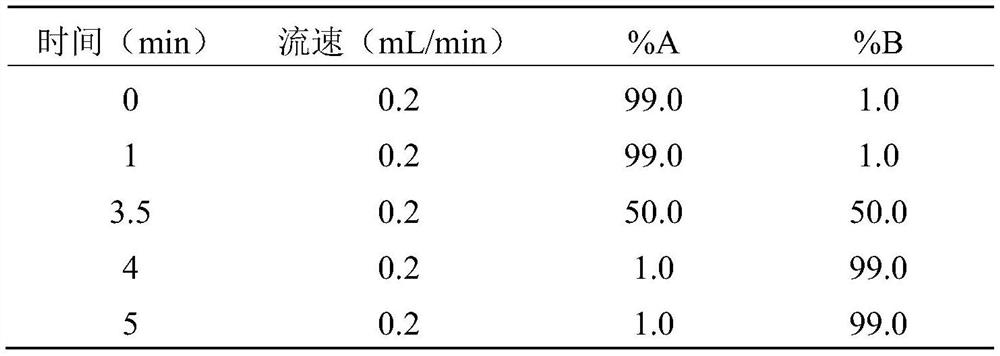
[0044] (2) Design primer sequences to amplify the cysteine transporter gene from the Escherichia coli genomic DNA obtained in the above step (1), the amplification conditions are as follows: pre-denaturation at 94°C for 3min, followed by 30 cycles of 94°C 20s, 55°C for 20s, 72°C for 3min, and finally 72°C for 10min; the specific primers are as follows:
[0045] The gene encoding cysteine transporter Bcr: utilize primer Bcr-F (nucleotide sequence as shown in SEQ ID NO.15) and Bcr-R (nucleotide sequence as shown in SEQ ID NO.16) from step Amplify the Bcr gene in the Escherichia coli genomic DNA obtained in (1)...
Embodiment 3
[0051] Example 3 Construction of recombinant Bacillus subtilis / pP43NMK-SMT / pSTOP1622-cysteine transporter
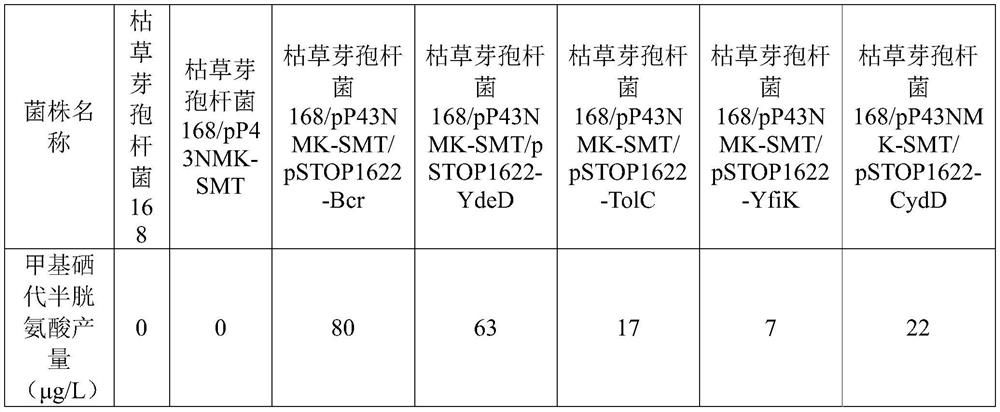
[0052] The expression vectors pSTOP1622-Bcr, pSTOP1622-YdeD, pSTOP1622-TolC, pSTOP1622-YfiK, pSTOP1622-CydD were transformed into recombinant Bacillus subtilis / pP43NMK-SMT, respectively, to obtain recombinant Bacillus subtilis / pP43NMK-SMT / pSTOP1622-Bcr, Recombinant Bacillus subtilis / pP43NMK-SMT / pSTOP1622-YdeD, Recombinant Bacillus subtilis / pP43NMK-SMT / pSTOP1622-TolC, Recombinant Bacillus subtilis / pP43NMK-SMT / pSTOP1622-YfiK, Recombinant Bacillus subtilis / pP43NMK-SMT / pSTOP1622- CydD; the above transformation products were respectively cultured on kanamycin and tetracycline-resistant plates, and cultured upside down at 37°C for 12 hours until colonies appeared, and a single colony was picked and cultured for preservation, and the genome was extracted for PCR identification, and the recombinants were obtained respectively. Recombinant Bacillus subtilis / pP43NMK-SMT / pSTOP162...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com