Recombinant engineering bacterium and application thereof in preparation of generic compounds
A technology for recombining engineering bacteria and recombinant plasmids, applied in the field of genetic engineering, can solve the problems of limiting industrial application prospects, cumbersome reaction steps, and long reaction time, and achieve the effects of shortening the production cycle, low reaction cost, and avoiding cumbersome steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
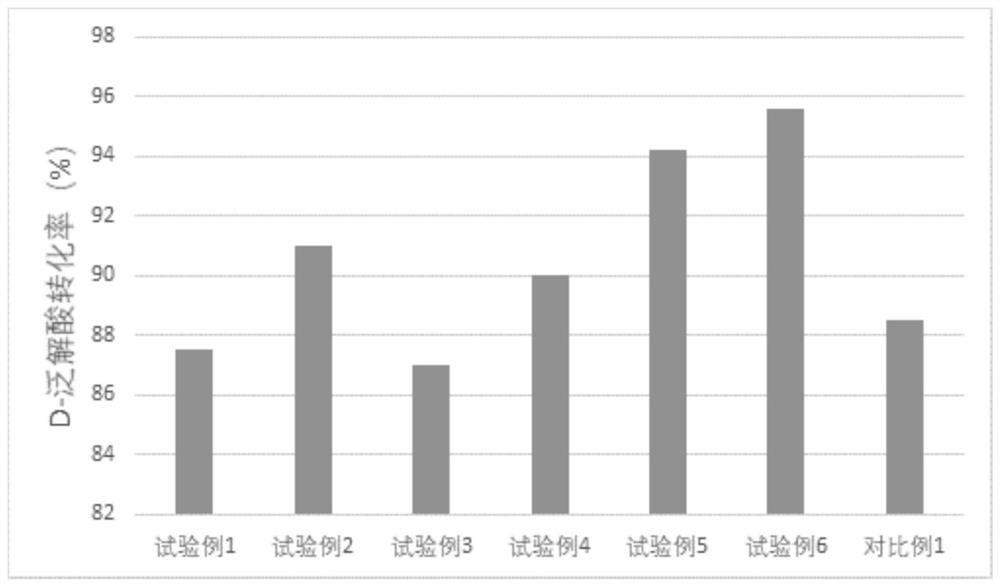
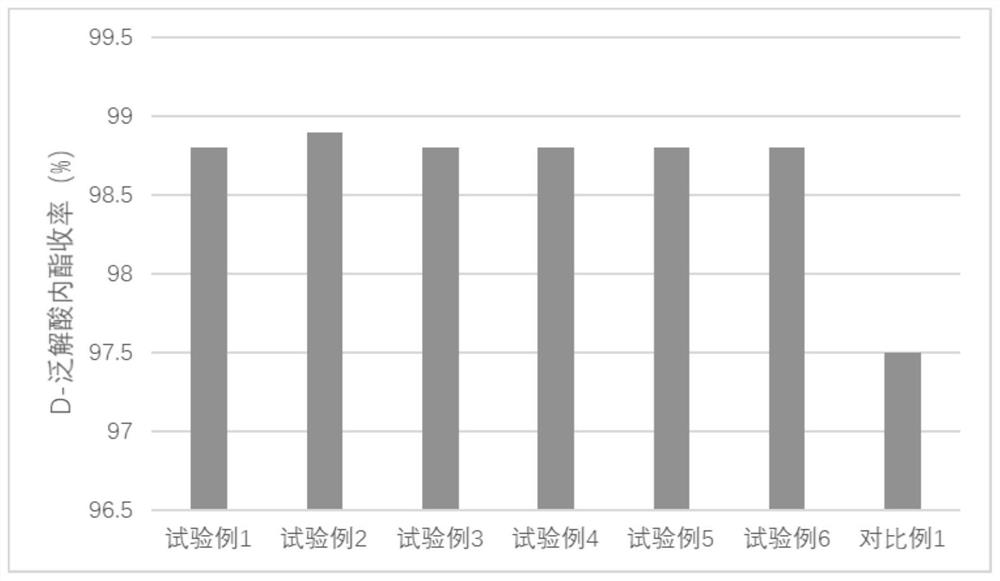
Examples
Embodiment 1
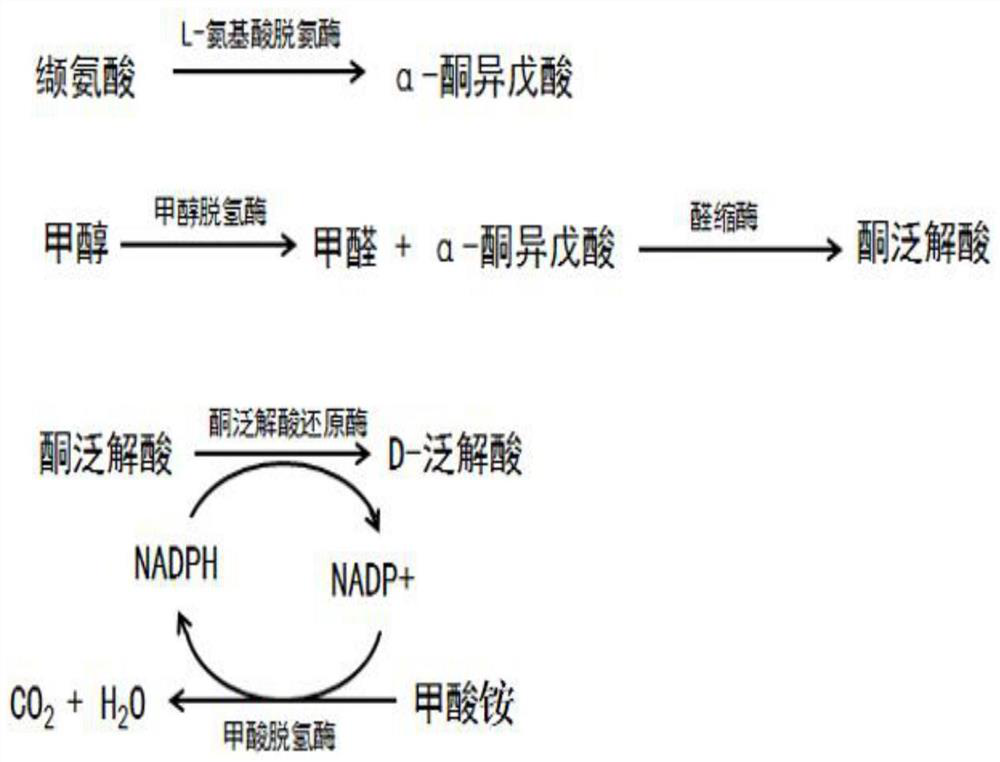
[0121] Example 1 Construction of the first recombinant engineering bacteria expressing L-amino acid deaminase
[0122] Step 1, according to the nucleotide sequence of the L-amino acid deaminase encoding gene of Proteus mirabilis (the encoded amino acid sequence is shown in SEQ ID NO: 6) according to Escherichia coli (E.coli) The codon preference of L-amino acid deaminase is optimized, and the L-amino acid deaminase optimized gene sequence is obtained, and its nucleotide sequence is shown in SEQ ID NO: 1;
[0123] The nucleotide sequence shown in SEQ ID NO: 1 is artificially synthesized, and XhoI and NdeI restriction sites are added to obtain the target gene 1.
[0124] Step 2, using the DNA molecule of the target gene 1 as a template, using primer pairs LA-for and LA-rev to carry out PCR amplification, 1% agarose gel electrophoresis to separate the PCR products, and then use a gel recovery kit to recover the target gene 1. gene fragments.
[0125] The primer sequences are ...
Embodiment 2
[0134] Example 2 Construction of the second recombinant engineering bacteria co-expressing methanol dehydrogenase and aldolase
[0135] Step 1, according to the nucleotide sequence of the methanol dehydrogenase encoding gene sequence of Bacillus methanolicus (the encoded amino acid sequence is shown in SEQ ID NO: 7) according to the code of Escherichia coli (E.coli) Codon optimization was carried out according to the sub-preference to obtain the methanol dehydrogenase optimized gene sequence, and its nucleotide sequence is shown in SEQ ID NO: 2;
[0136] The nucleotide sequence of the aldolase-encoding gene sequence derived from Escherichia coli (the encoded amino acid sequence is shown in SEQ ID NO: 8) was determined according to the codon bias of Escherichia coli. Codon optimization, the aldolase optimization gene sequence is obtained, and its nucleotide sequence is as shown in SEQ ID NO:3;
[0137] The nucleotide sequences shown in SEQ ID NO: 2 and SEQ ID NO: 3 were arti...
Embodiment 4
[0156] Example 4Construction of the third recombinant engineering bacteria co-expressing formate dehydrogenase and ketopantoate reductase
[0157] In step 1, the nucleotide sequence of the gene sequence encoding formate dehydrogenase from Burkholderia stabilis (the encoded amino acid sequence is shown in SEQ ID NO: 9) is based on Escherichia coli (E. .coli) codon preference carries out codon optimization, and adds BamHI and NotI restriction enzyme cleavage site, obtains formate dehydrogenase optimization gene sequence, and its nucleotide sequence is as shown in SEQ ID NO:4;
[0158] The nucleotide sequence of the ketopantoate reductase-encoding gene sequence (the encoded amino acid sequence thereof is shown in SEQ ID NO: 10) derived from Stenotrophomonas maltophilia was based on Escherichia coli (E. coli), the codon preference is optimized, and XhoI and NdeI restriction sites are added to obtain a ketopantoate reductase-optimized gene sequence, whose nucleotide sequence is s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com