SILAC-based mass spectrum method for absolute quantification of serum cystatin C
An absolute quantitative and cystatin technology, applied in the field of mass spectrometry detection, can solve problems such as inaccurate quantitative results, achieve the effect of improving sensitivity, anti-interference ability, and high extraction efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 115
[0033] Example 1 15 Prokaryotic expression and protein purification of recombinant cystatin C labeled with N isotope
[0034] 1) Target gene synthesis
[0035] The amino acid sequence of Cys C protein was retrieved from the Pubmed protein library, and the sequence was optimized. The amino acids 1-20 of the analyzed protein are the transmembrane region, then the signal peptide is removed, and a 6xHis tag is fused to the carboxyl terminus of the sequence. The purpose is to facilitate the purification of proteins using nickel columns in subsequent experiments. The corresponding sequence was added during optimization, and the molecular weight of the target protein was about 15kD after optimization. The corresponding nucleic acid sequences were obtained in the Pubmed gene bank. For the sequence analysis, designs such as codon usage preference adjustment, codon usage distribution optimization and GC content optimization were performed in PrimerPrimer 5.0 software. Analysis of r...
Embodiment 2
[0075] Example 2 Selection of Cys C Featured Peptides
[0076] (1) Cys C protein sequence information
[0077] The Cys C protein sequence information from the NCBI protein database is as follows:
[0078] Cystatin-C precursor[Homo sapiens]
[0079] NCBI Reference Sequence: NP_000090.1
[0080] The protein sequence is as follows:
[0081] 1 magplrapllllailavalavspaagsspgkpprlvggpmdasveeegvrraldfavgey
[0082] 61 nkasndmyhsralqvvrarkqivagvnyfldvelgrttctktqpnldncpfhdqphlkrk
[0083] 121 afcsfqiyavpwqgtmtlskstcqda (SEQ ID NO: 1)
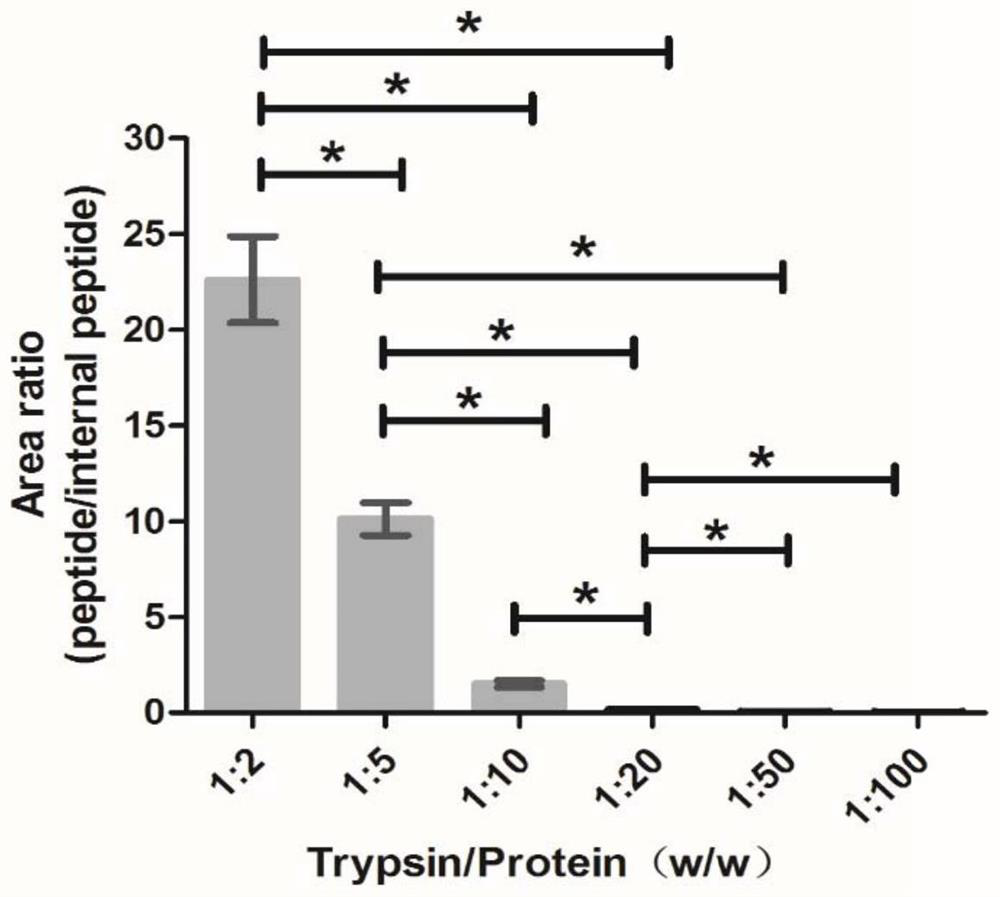
[0084] (2) Prediction of peptides produced by Cys C hydrolysis by trypsin.
[0085] The PeptideMass tool in the ExPASy software predicts the peptide segment produced by Cys C hydrolysis by trypsin, the theoretical isoelectric point pI: 8.75 / Mw (average mass), and the molecular weight: 13347.14.
[0086] (3) Retrieval and analysis of physicochemical properties of characteristic peptides.
[0087] The physicochemical properties of the characteristi...
Embodiment 3
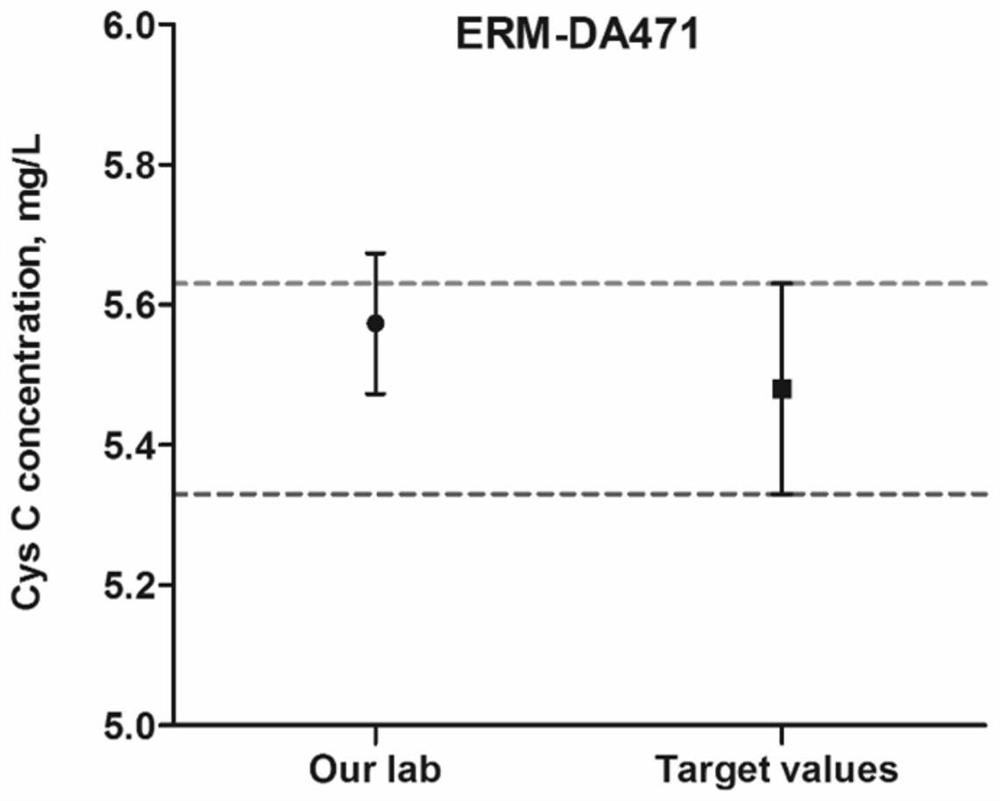
[0089] Example 3 Determination of Mass Spectrometry Conditions
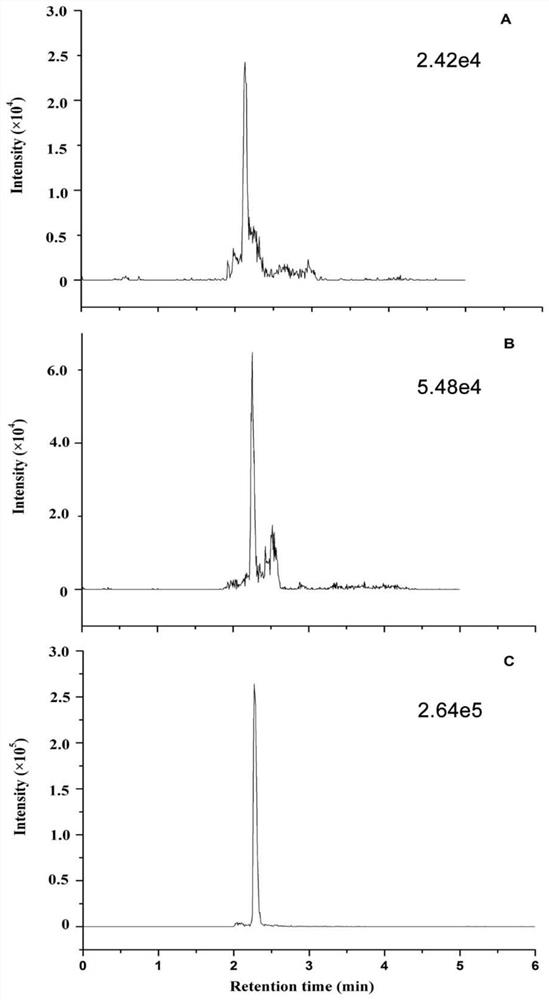
[0090] Confirmation of peptide standard mass spectrometry conditions: For the screened characteristic peptides, the biological company will synthesize the characteristic peptides and their isotope-labeled internal standards. Prepare 1 mL of standard peptides with a concentration of 1 μg / mL, and inject by needle pump.
[0091] Cys C protein standard and 15 N-labeled Cys C protein standard mass spectrometry confirmation: 10 μg Cys C or 15 The N-labeled Cys C protein standard was subjected to protein denaturation, reductive alkylation and proteolysis according to the protein sample pretreatment steps, and then injected by needle pump.
[0092] (1) Ion pair selection
[0093] 1) Q1 MS first-level full scan.
[0094] 2) Characteristic peptide product ion scan (Product Ion MS 2)
[0095] Enter the mass-to-charge ratio of the precursor ion;
[0096] Optimize CE, range 5-30, observe the signal intensity of product ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com