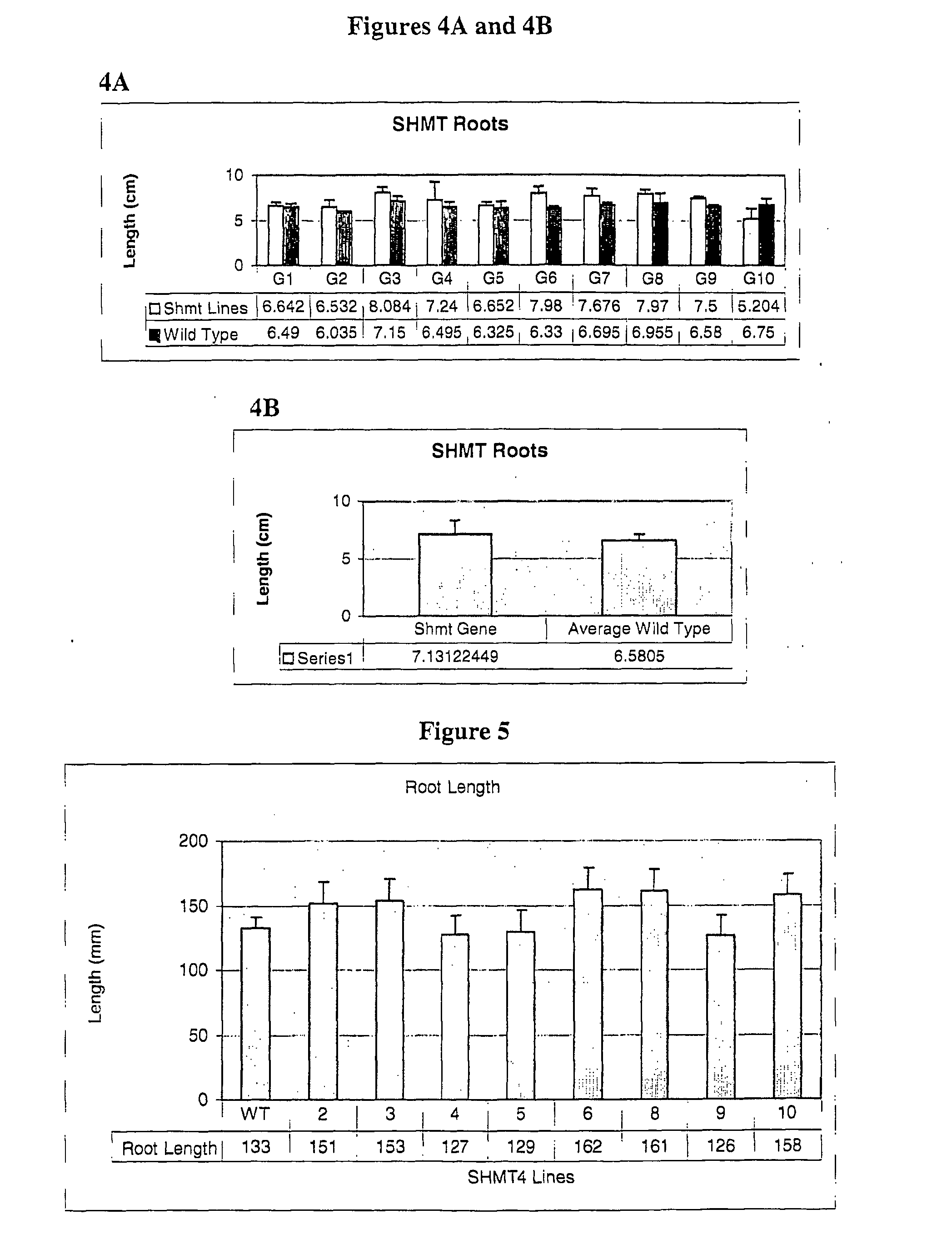
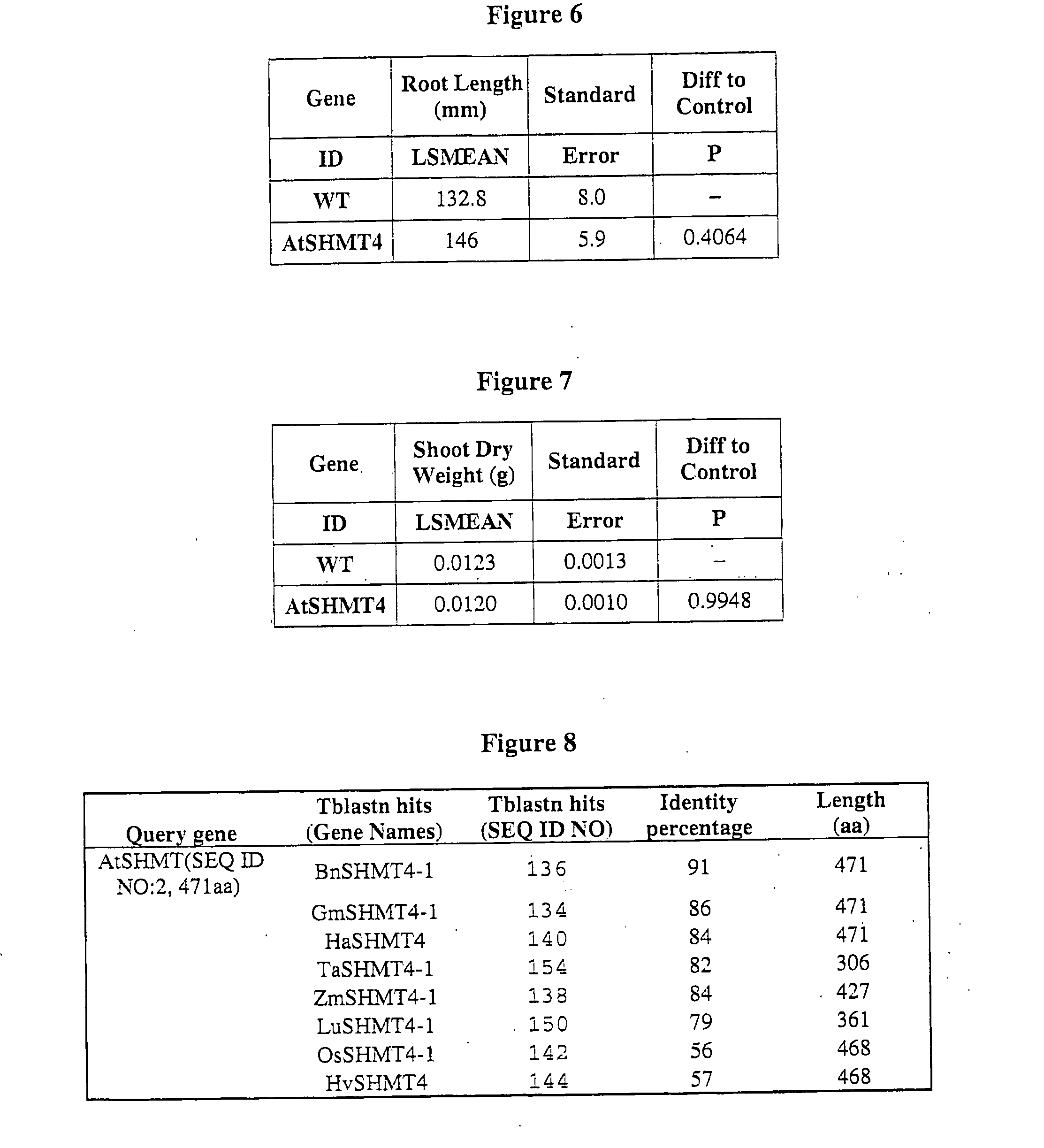
Yield Increase in Plants Overexpressing the SHSRP Genes
a technology of shsrp and plant, applied in the field of nucleic acid sequences encoding polypeptides, can solve the problems of limiting the growth and productivity of plants, restricting the use of controlled environments for testing yield differences, and profound effects on the development, growth, plant size, and yield of most crop plants. , to achieve the effect of increasing tolerance to environmental conditions, increasing root growth, and increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Total DNA Isolation from Plant Material
[0175]The details for the isolation of total DNA relate to the working up of one gram fresh weight of plant material. The materials used include the following buffers: CTAB buffer: 2% (w / v) N-cethyl-N,N,N-trimethylammonium bromide (CTAB); 100 mM Tris HCl pH 8.0; 1.4 M NaCl; 20 mM EDTA; N-Laurylsarcosine buffer: 10% (w / v) N-laurylsarcosine; 100 mM Tris HCl pH 8.0; and 20 mM EDTA.
[0176]The plant material was triturated under liquid nitrogen in a mortar to give a fine powder and transferred to 2 ml Eppendorf vessels. The frozen plant material was then covered with a layer of 1 ml of decomposition buffer (1 ml CTAB buffer, 100 μl of N-laurylsarcosine buffer, 20 μl of β-mercaptoethanol, and 10 μl of proteinase K solution, 10 mg / ml) and incubated at 60° C. for one hour with continuous shaking. The homogenate obtained was distributed into two Eppendorf vessels (2 ml) and extracted twice by shaking with the same volume of chloroform / isoamyl alcohol (24...
example 2
Isolation of Total RNA and cDNA from Arabidopsis Plant Material
[0177]AtSHMT4 was isolated by preparing RNA from Arabidopsis leaves using the RNA mini-isolation kit (Qiagen kit) following the manufacturer's recommendations. Reverse transcription reactions and amplification of the cDNA were performed as described below.[0178]1. Use 2 μl of RNA (0.5-2.0 μg) preparation in a 10 μl Dnase reaction, move the tube to 37° C. for 15 minutes, add 1 μl 25 mM EDTA, and then heat reaction to 65° C. for 15 minutes.[0179]a. Buffer (10×: 200 mM Tris, 500 mM KCl, 20 mM MgCl2)—1 μl[0180]b. RNA—2 μl[0181]c. Dnase (10 U / μl)−1 μl[0182]d. H2O—6 μl[0183]2. Use 1 μl of the above reaction in a room temperature reaction, and heat to 65° C. for 5 minutes.[0184]a. Dnased RNA (0.025-0.1 μg depending on the starting amount)—1 μl[0185]b. 10 mM dNTPs—1 μl[0186]c. Primer (10 μM)—1 μl[0187]d. H2O—up to 1091[0188]3. Prepare a reaction mix with these reagents in a separate tube[0189]a. SuperScript II RT buffer (10×)—2 ...
example 3
Cloning of AtSHMT4
[0200]The cDNA isolated as described in Example 2 was used to clone the AtSHMT4 gene by RT-PCR. The following primers were used: The forward primer was 5′-ATGGAACCAGTCTCTTCATG-3′ (SEQ ID NO:159). The reverse primer was 5′-CTAATCCTTGTACTTCATCT-3′ (SEQ ID NO:160). PCR reactions for the amplification included: 1×PCR buffer, 0.2 rmLM dNTP, 100 ng Arabidopsis thaliana DNA, 25 pmol reverse primer, 2.5 u Pfu or Herculase DNA polymerase.
[0201]PCR was performed according to standard conditions and to manufacturer's protocols (Sambrook et al., 1989, Molecular Cloning, A Laboratory Manual, 2nd Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., Biometra T3 Thermocycler). The parameters for the reaction were: 1 cycle for 3 minutes at 94° C.; followed by 25 cycles of 30 seconds at 94° C., 30 seconds at 55° C., and 1.5 minutes at 72° C.
[0202]The amplified fragments were then extracted from agarose gel with a QIAquick Gel Extraction Kit (Qiagen) and ligated in...
PUM
| Property | Measurement | Unit |
|---|---|---|
| weight | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com