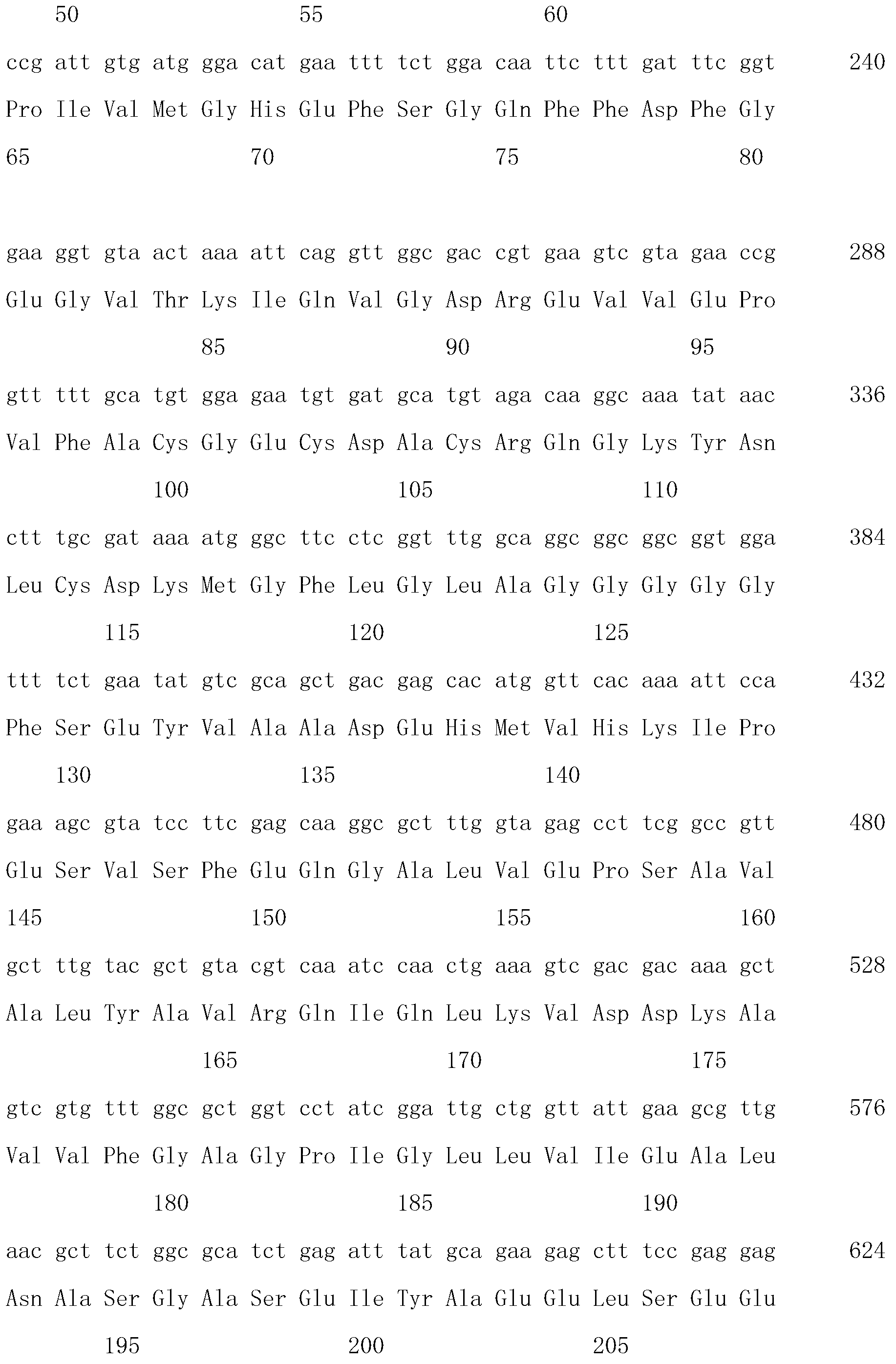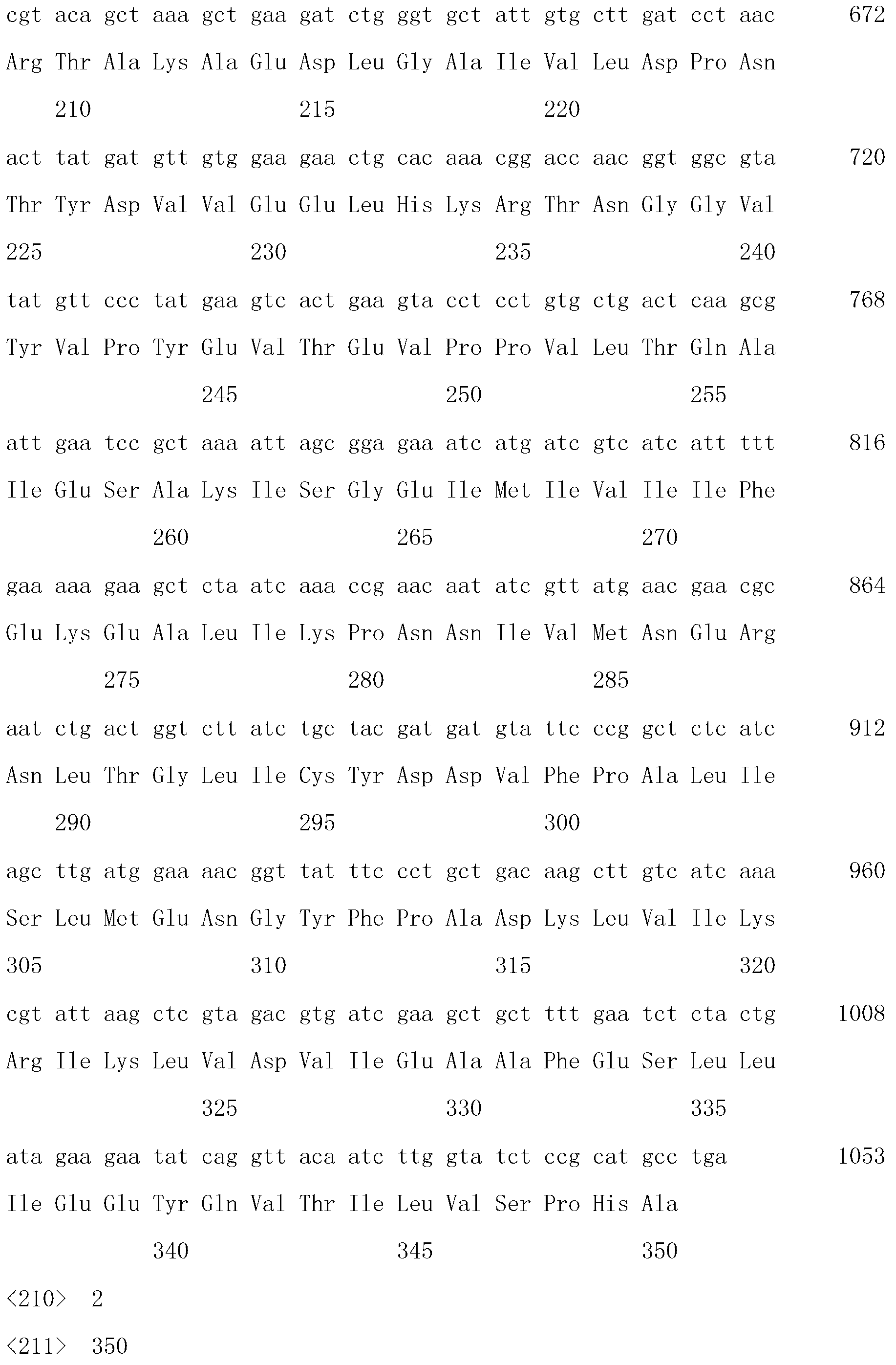Genetically engineering bacterium capable of producing S-3-hydroxy-butanone as well as construction method and application thereof
A technology of genetically engineered bacteria and hydroxybutanone is applied in the field of genetically engineered bacteria and its construction, and can solve the problems of high price of meso-2,3-butanediol, serious environmental pollution, and many reaction steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] The preparation method of Jerusalem artichoke inulin crude extract:
[0039] Fresh Jerusalem artichoke is washed and peeled, blanched to eliminate enzymes (100°C, 15min), cut into thin slices, blown and dried at 70°C, and then crushed through an 80-mesh sieve to obtain coarse powder of Jerusalem artichoke, which is stored in the refrigerator for later use. According to the ratio of 1:6, weigh the coarse powder of Jerusalem artichoke into the water, stir evenly, heat and extract in a water bath at 70°C for 4 hours, adjust the pH to 9 with milk of lime, then keep it warm in a water bath at 80°C for 1 hour, and filter it with gauze to obtain the Jerusalem artichoke. Crude powder extract.
[0040] Embodiment: 2: the cloning of gene Bud A:
[0041] The primers required for synthetic PCR were designed according to the sequence of the Bud A gene in Paenibacillus polymyxa ATCC 12321 published by Genebank:
[0042] P1: 5'-ATGCAAGCATTGAGATGGCA-3'
[0043] P2: 5'-TTAGGCTTTCGGAG...
Embodiment 3
[0045] Example 3: Recombinant cosmid "SuperCos-1 / pIJ790-Bud A homology arm Ⅰ-oriT-Apra R - Construction of Bud A homology arm Ⅱ":
[0046] Introduce EcoR I and Xho I at both ends of the 39bp base sequence at the 5' end of Bud A to obtain the homology arm I of Bud A; introduce Not I at both ends of the approximately 20bp base sequence at the 3' end of Bud A , BamH I, to get the Bud A homology arm II. The oriT replication initiation sequence and the apramycin resistance gene Apramycin were connected to the clone cosmid SuperCos-1 / pIJ790 according to the restriction site R The two ends of the sequence were obtained containing "Bud A homology arm Ⅰ-oriT-Apra R - The recombinant cosmid SuperCos-1 / pIJ790 of the Bud A homology arm II" sequence and the λRed sequence.
Embodiment 4
[0047] Example 4: Recombinant cosmid "SuperCos-1 / pIJ790-Bud A homology arm Ⅰ-oriT-Apra R -Bud A Homology Arm II" was transformed into Paenibacillus polymyxa CGMCC 3044.
[0048] The recombinant cosmid SuperCos-1 / pIJ790-"SuperCos-1 / pIJ790-Bud A homology arm Ⅰ-oriT-Apra R -Bud A Homologous Arm II" was transformed into Paenibacillus polymyxa polymyxa CGMCC 3044, spread on a plate containing 40 μg / mL apramycin, picked positive recombinants, and carried out colony PCR identification to obtain recombinant Paenibacillus polymyxa Bacillus named paenibacillus polymyxa CGMCC 3044-Bud A - .
PUM
| Property | Measurement | Unit |
|---|---|---|
| optical purity | aaaaa | aaaaa |
| optical purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com