Heat-resisting glucamylase as well as coding gene and application thereof
A technology of glucoamylase and coding gene, which is applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problems of Mucor micromycetes that have not been reported, and achieve the effect of good thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1. Amplification of Glucoamylase Gene
[0024] 1.1 Strain and its culture
[0025] Rhizomucor pusillus (strain number 3.204) was purchased from CGMCC, General Microbiology Center of China Microorganism Culture Collection and Management Committee in Beijing.
[0026] Mucor spores were washed from PDA slants cultured for 5-6 days with sterile water, inoculated with liquid medium, and cultured at 30°C for 2-3 days to collect mycelia for genome or RNA extraction.
[0027] 1.2 Genome extraction
[0028] Refer to Omega Fungal Genome Extraction Kit Instructions
[0029] 1.3 Total RNA extraction
[0030] The spores on the slanted surface of the PDA cultured for 6 days were washed with sterile water, inoculated with a fungal amylase-producing liquid medium, cultured for 2-3 days, and the mycelia were collected by gauze filtration. Follow the instructions of Takara RNA extraction reagent.
[0031] 1.4 cDNA first-strand synthesis
[0032] Using the extracted total mR...
Embodiment 2
[0093] Example 2. Construction and transformation of Pichia pastoris recombinant expression vector containing glucoamylase gene
[0094] 2.1 Primer design
[0095] The sequence of R.pGEcoRf is SEQ ID NO. 23: GGAATTCATGCGTTATGCAACCCCGC
[0096] The sequence of R.pGNotr is SEQ ID NO. 24: TTGCGGCCGCTTACCCTCTTTTGACCA
[0097] 2.2 Construction of recombinant expression vector
[0098] Using the pMD19-T Vector with the open reading frame of the glucoamylase gene as a template, PCR amplification was carried out with the primers shown in 3.1; the PCR products were digested with EcoRI and NotI, and then expressed with the yeast that was also digested with EcoRI and NotI. The vector pPIC9k was ligated and screened to obtain a recombinant plasmid (pPIC9KR.pGLA).
[0099] 2.3 Preparation of Pichia pastoris competent cells
[0100] (1) Inoculate yeast in YPD liquid medium and cultivate at 30°C to OD1-2A (600nm)
[0101] (2) Centrifuge the culture solution at 6000rpm at 4°C for 3min, a...
Embodiment 3
[0111] Example 3. Inducible expression of Pichia pastoris
[0112] 3.1 Pick a single clone of the recombinant bacteria on the G418-containing YPD plate described in 3.5, inoculate the YPD liquid medium, and cultivate at 30°C and 200rpm for 24h.
[0113] 3.2 The above-mentioned seed culture medium was inserted into BMGY medium, and cultured at 30°C and 200rpm for 24h.
[0114] 3.3 Culture to OD6.0, 6500rpm, centrifuge for 5min to collect bacterial cells, transfer to BMMY medium, 225rpm, and induce expression at 30°C, add 100% methanol every 24h to a final concentration of 0.5%.
[0115] 3.4 After induction for 120 h, the supernatant was collected by centrifugation, the enzyme activity was measured, the protein concentration was measured, and protein electrophoresis was performed.
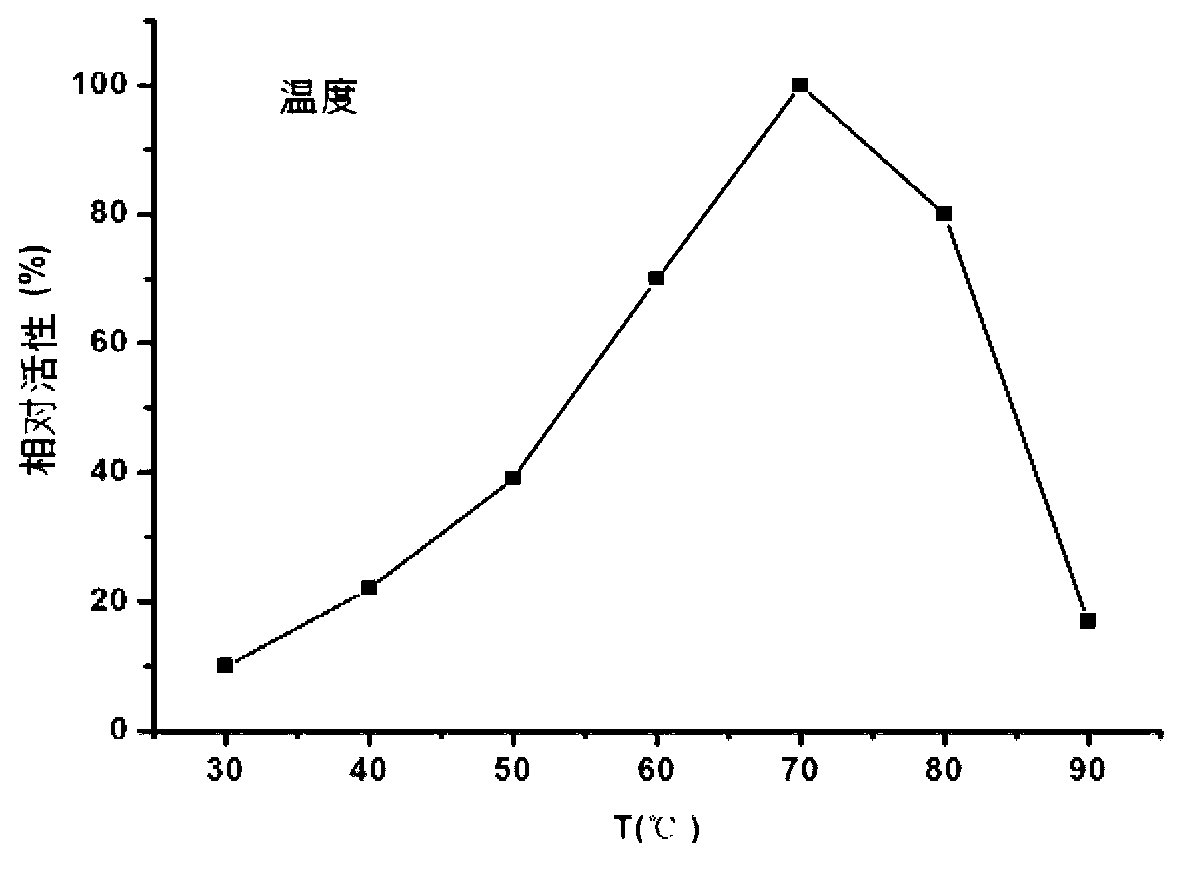
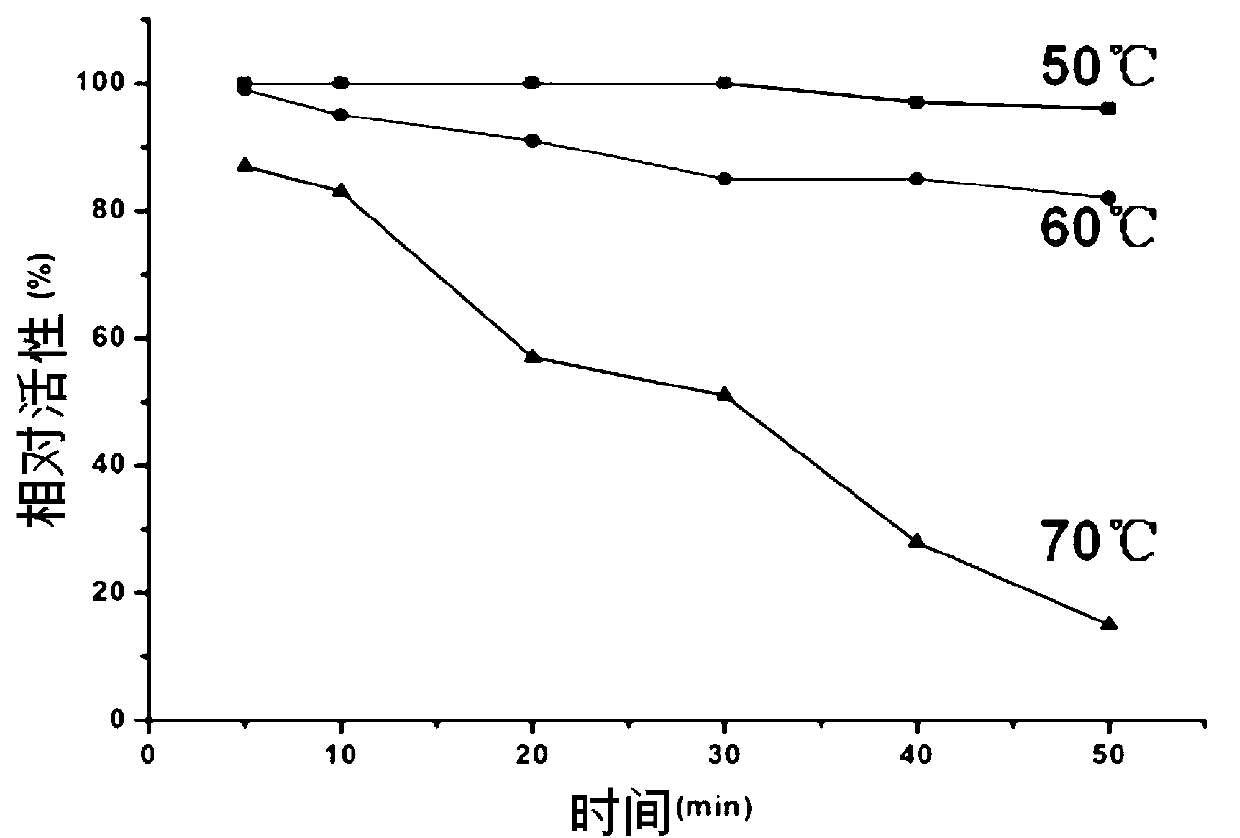
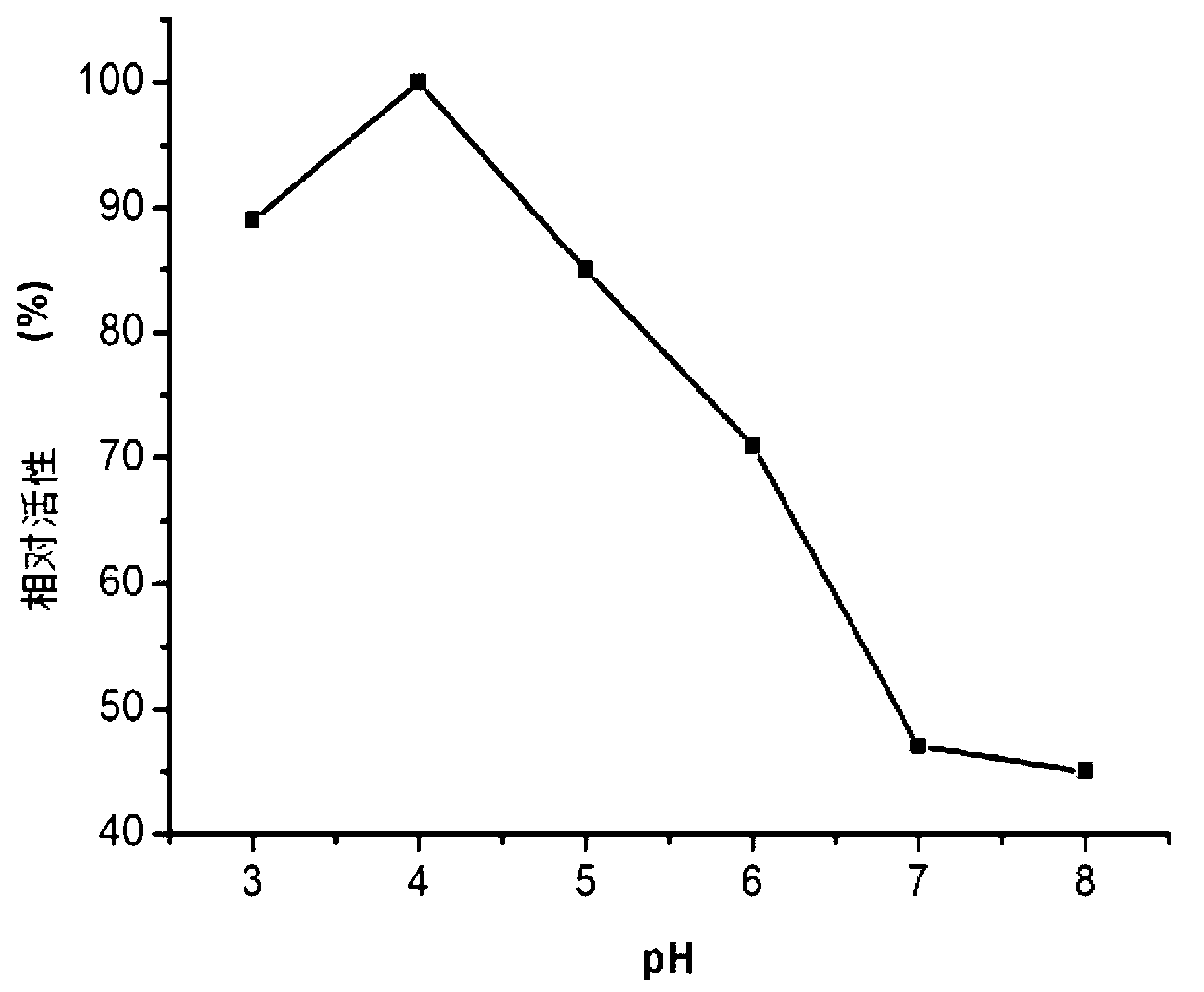
[0116] figure 1 The electropherogram showing the induced expression of Mucor glucoamylase Pichia pastoris KM71 recombinant bacteria.
[0117] The recombinant Pichia pastoris constructed by the pre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com