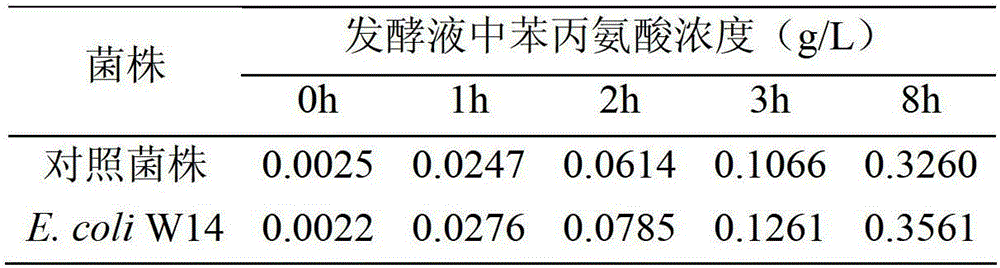
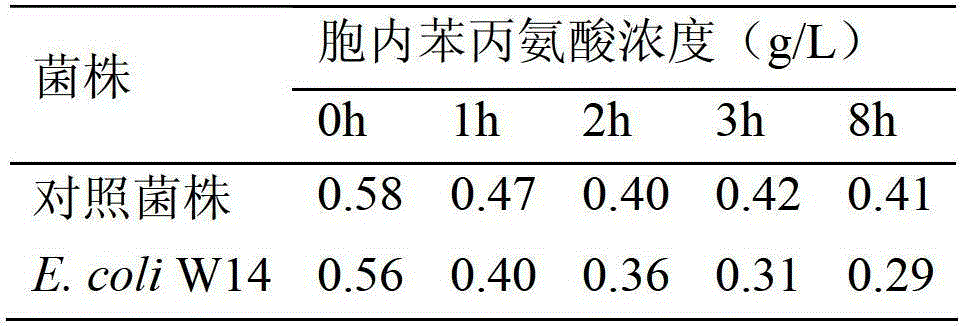
Method for enhancing L-phenylalanine exocytosis of escherichia coli
A technology of phenylalanine cells and Escherichia coli, which is applied in the field of bioengineering, can solve the problems of mutant strains with nutritional deficiencies, limited amino acid accumulation, and difficult to release regulatory sites, so as to improve fermentation intensity and enhance the level of extracellular secretion , Accelerate the effect of extracellular transport rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Construction of the plasmid pR15ABK overexpressing AroG, PheA, AroK and YdiB
[0024] 1.1 Release of metabolic regulation of 3-deoxy-D-arabinoheptulose-7-phosphate synthase AroG
[0025]Using the fitted DNA of Escherichia coli as a template, the upstream primer aroG15_F146 (base sequence shown in SEQ ID NO: 2) and the downstream primer aroG_HindⅢ_RV (base sequence shown in SEQ ID NO: 3) were used for PCR amplification to obtain gene fragments A, using Escherichia coli nucleoid DNA as a template, PCR amplification was performed with upstream primer aroG_Kpn Ⅰ_FW (base sequence shown in SEQ ID NO:4) and downstream primer aroG15_R146 (base sequence shown in SEQ ID NO:5) Obtain gene fragment B.
[0026] Using gene fragments A and B as templates, by overlapping PCR (according to the PCR site-directed mutagenesis technique, the references are as follows: Ho, S.N., H.D. Hunt, R.M. Horton, J.K. Pullen, and L.R. Pease (1989) Site-directed mutagenesis by overlap extens...
Embodiment 2
[0032] Example 2 Release of metabolic regulation of tyrosine transaminase TyrB
[0033] The tyrB coding gene was obtained by PCR amplification using the fitted DNA of Escherichia coli as a template, and the primers were designed as follows: the base sequence of the upstream primer tyrB_EcoRV_SD_SB_FW is shown in SEQ ID NO:14, and the base sequence of the downstream primer tyrB_AflⅡ_EcoRI_RV is shown in SEQ ID NO:15 , wherein, the SD sequence AAGGAGGAACAGAC is the ribosome binding site, and the SB sequence ATGTTCCAGAAGGTCGATG (as shown in SEQ ID NO: 1) is the starting base sequence of the coding frame of the tyrB gene after replacement.
[0034] The amplified TyrB coding gene was connected to the pMD 18T-simple Vector, sequenced and identified correctly by Shanghai Sangon Bioengineering Co., Ltd., and then ligated by enzyme digestion (for specific methods, please refer to the corresponding restriction endonuclease or ligase instructions) and The plasmid pR15ABK constructed in E...
Embodiment 3
[0040] Example 3 Release of metabolic regulation of methyl viologen efflux pump (YddG)
[0041] The yddG coding gene was obtained by PCR amplification using E. coli fitting DNA as a template, and the primers were designed as follows: the base sequence of the upstream primer yddG_EcoRV_SD_FW is shown in SEQ ID NO:16, and the base sequence of the downstream primer yddG_NcoI_KpnI_RV is shown in SEQ ID NO:17 , wherein, the SD sequence AAGGAGGAACAGAC is the ribosome binding site.
[0042] Ligate the amplified yddG-encoding gene to pMD 18T-simple Vector, after sequenced and identified correctly by Shanghai Sangon Bioengineering Co., Ltd., carry out an enzyme digestion ligation reaction (see the corresponding restriction enzyme or ligase instructions for specific methods) Connect with the plasmid constructed in Example 2, place the yddG coding gene in the strong promoter P L Later, a recombinant expression plasmid that can overexpress YddG, TyrB, AroG, PheA, AroK and YdiB at the sam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com