Malus hupehensis MhWRKY40a gene and applications thereof
A gene, crabapple technology, applied to the 'Hubei Begonia' MhWRKY40a gene and its application fields, can solve the problems of reducing production costs, squeezing living space, and threatening grain and oil security.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] RNA was extracted from leaves of Begonia hubei by CTAB method, and then the RNA was reverse-transcribed into cDNA, and the target gene was cloned from the cDNA template of leaves by RT‐PCR. PCR reaction system is template cDNA 1μL (50ng·μL ‐1 ), 10×PCR Buffer2.5μL, MgCl 2 1.5μL (25mmol·L ‐1 ), dNTP2.0μL (2.5mmol L ‐1 ), upstream primer F1 (SEQ ID NO.2), downstream primer R1 (SEQ ID NO.3) each 1.0 μL (100ng·μL ‐1 ), 1.0U rTaq enzyme, made up to 25 μL with ultrapure water. The PCR program was 94°C for 4min; 94°C for 30s, 56°C for 30s, 72°C for 2min, 35 cycles; 72°C for 10min. Take 20 μL of the PCR product and analyze it by electrophoresis on a 1.0% agarose gel. After the target fragment was recovered and connected to the pMD19-TSimple (TaKaRa) vector, it was transformed into DH5α Escherichia coli, and more than 5 single colonies were randomly selected for sequencing. The gene sequence is shown in SEQ ID NO.1. After sequencing, the an...
Embodiment 2
[0031] Embodiment 2 vector construction
[0032] On the basis of restriction site analysis, BamHI and SacⅠ restriction sites were introduced at the 5′ end of the original gene cloning primer, and primers with restriction sites (underlined partial sequence) were designed, namely the upstream primer K-MhWRKY40a F (SEQ ID NO.4), downstream primer K‐MhWRKY40a R (SEQ ID NO.5). Using the correctly sequenced returned plasmid in Example 1 as a template, a gene fragment with a restriction site was amplified by PCR. The reaction system is as follows:
[0033]
[0034]The PCR program was 94°C for 4min; 94°C for 120s, 65°C for 30s, 72°C for 2min, 35 cycles; 72°C for 10min. All PCR products were analyzed by 1.0% agarose gel electrophoresis. The target fragment was recovered and connected to PMD19-T Simple, the ligated product was transformed into DH5α Escherichia coli, and more than 5 single colonies were randomly selected for sequencing. The recovery fragment connection system is a...
Embodiment 3
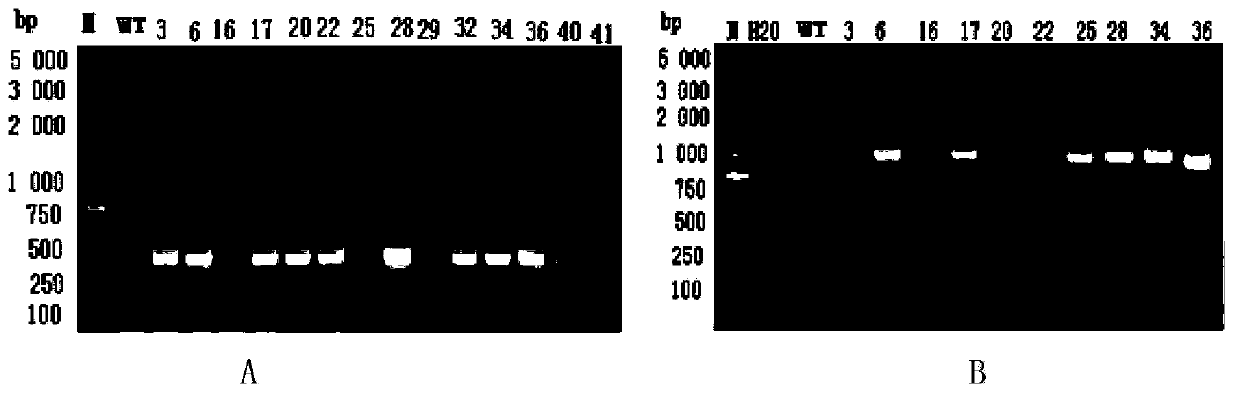
[0054] Embodiment 3 MhWRKY40a Gene Transformation Tobacco and Detection
[0055] The transformation process requires no bacterial contamination and is carried out on an ultra-clean workbench. Inoculate a single colony of transformed Agrobacterium EHA105 in a solution containing 50ug·mL ‐1 Rif, 50ug·mL ‐1 Shake culture in Km’s YEB liquid medium at 28°C and 250rpm until OD600 is about 0.5-1.0, divide into 50mL centrifuges, fill each tube with 40mL, centrifuge at 5000rpm for 10min, discard the bacteria solution, and use 40ml of MS solution for precipitation (bacteria) Resuspend the culture medium, shake and activate at 28°C and 250rpm for 1h; cut tobacco leaves into 1-2cm 2 (leaf edges also need to be cut off to form wounds on the edges), put them into the activated Agrobacterium solution, soak for 3 to 5 minutes, and shake gently several times; take out the materials, put them on sterile filter paper to absorb excess bacteria solution, inoculated in the co-cultivation medium ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com