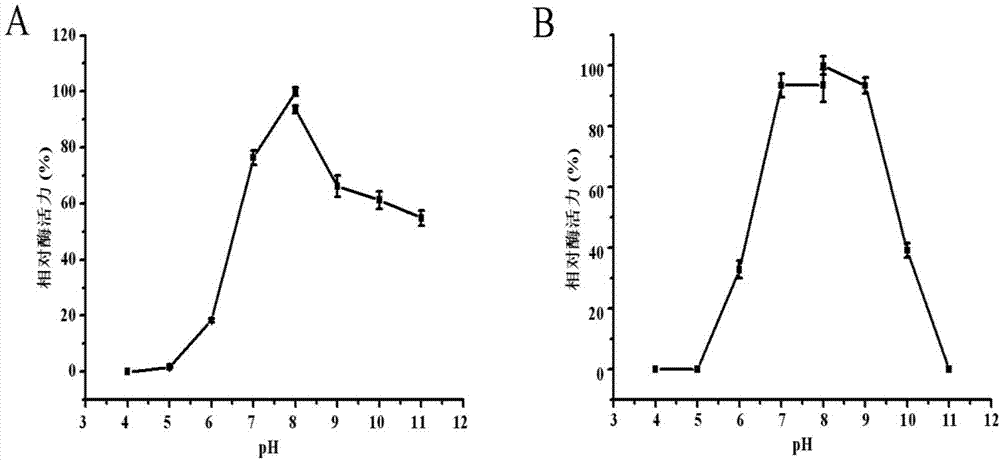
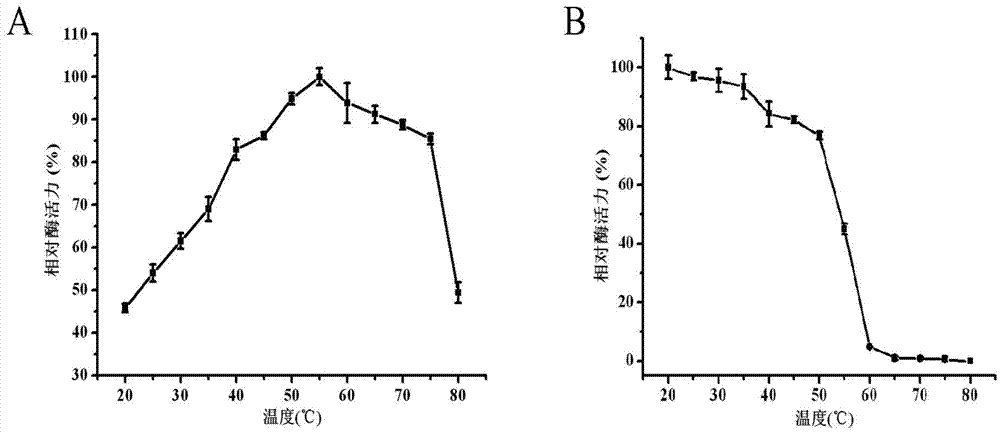
D-lactate oxidase and application thereof in D-lactic acid detection
A lactate oxidase and lactic acid technology, applied to lactate oxidase and its application fields, achieves the effects of easy fixation, good stability and good application potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1: Construction of Escherichia coli recombinant expression strain comprising Gluconobacter oxidans 621H D-lactate oxidase gene
[0028] 1. PCR amplification of GOX2071 gene of Gluconobacter oxidans 621H D-lactate oxidase
[0029] Genomic DNA of Gluconobacter oxydans 621H is prepared by conventional methods. This process can refer to the method of small-scale preparation of bacterial genomes in the "Guide to Molecular Biology" published by Science Press to extract the genomic DNA of Gluconobacter oxydans 621H ;
[0030] Primers were designed to introduce the NdeI and XhoI restriction enzyme sites that can be inserted into the plasmid pET25b (Novagen). The primer sequences are as follows:
[0031] Upstream primer 5'-AAG CATATGCCGGAACCAGTCATGA-3', carrying an NdeI site;
[0032] Downstream primer 5'-CAA CTCGAG GCCCGTGTAAACAGCA-3', carrying an XhoI site.
[0033] To extract the genomic DNA of Gluconobacter oxydans 621H, PCR amplification was performed using t...
Embodiment 2
[0040] Example 2: Expression of Gluconobacter oxydans 621H recombinant D-lactate oxidase in Escherichia coli recombinant strain
[0041] Inoculate the glycerol tube of the recombinant strain Escherichia coli BL21 (pET25b-GOX2071) obtained in Example 1 into 5 mL of LB liquid medium containing 100 μg / mL ampicillin with a 1% inoculum size, and culture on a shaker at 37° C. for 6 to 12 hours Afterwards, transfer to 100mL LB liquid medium containing 100μg / mL ampicillin with 1% inoculum amount, culture on a shaker at 37°C for 6-12 hours, and then transfer to 1L containing 100μg / mL ampicillin at 2.5% inoculum amount Penicillin LB liquid medium, cultured on a shaker at 37°C, when OD 600nm After reaching 0.4-0.8, IPTG with a final concentration of 1 mmol / L was added to induce expression, and the expression was induced at 37°C for 6 hours.
[0042] After the induction culture, the cells were collected by centrifugation at 6,000rpm for 10 minutes, and the cell pellet was washed twice wi...
Embodiment 3
[0043] Example 3: Purification of Gluconobacter oxydans 621H recombinant D-lactate oxidase
[0044] 1. Preliminary purification of D-lactate oxidase using Souce30Q anion exchange column
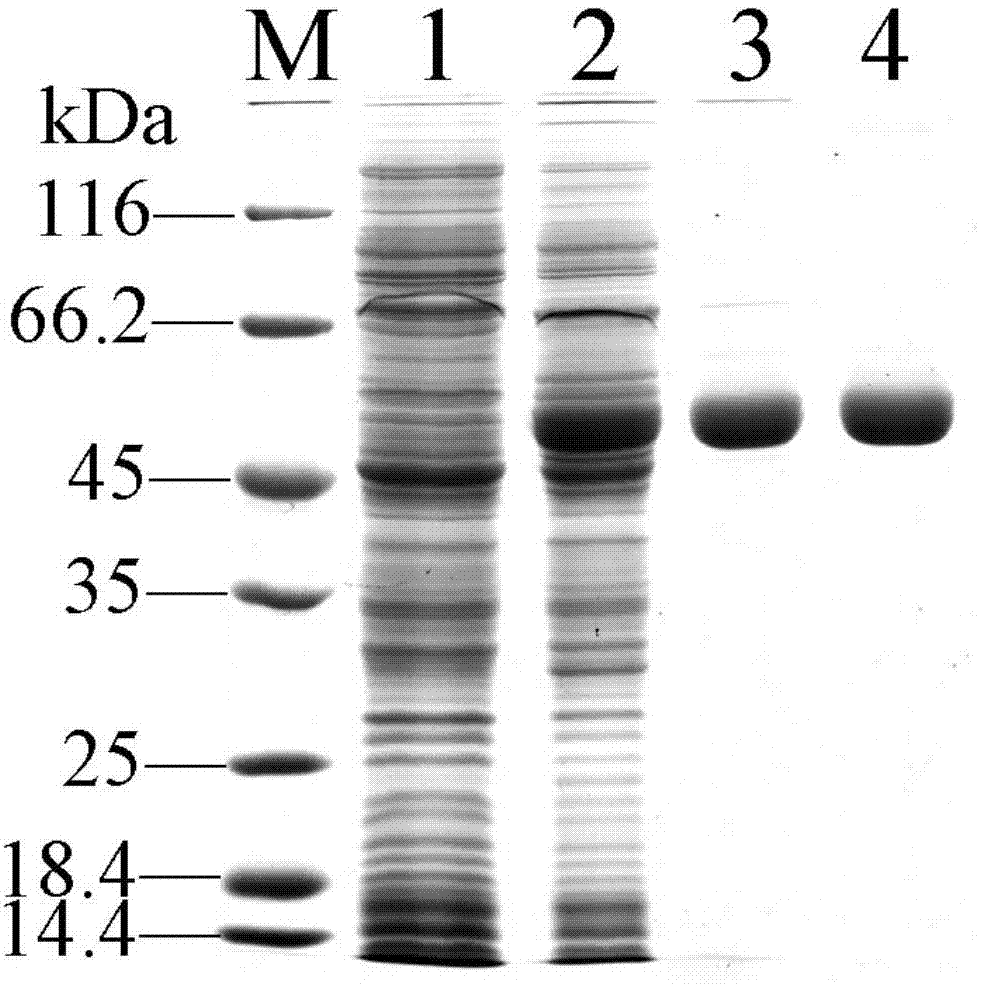
[0045] The crude enzyme liquid containing recombinant D-lactate oxidase obtained in Example 1 was filtered through a filter head with a pore size of 0.22 μm, and then placed on an anion exchange column Source30Q equilibrated with Buffer A, and an anion exchange column was used to elute with a gradient of Buffer B Elution, the elution gradient is 40%, 50%, 65%, 100%, collect the 65% gradient eluate containing D-lactate oxidase, promptly obtain the recombinant D-lactate oxidase ( figure 1 ).
[0046] The formula of the above anion exchange column elution buffer Buffer B is: 20mmol / L sodium phosphate, 500mmol / L sodium chloride, pH 7.4.
[0047] 2. Further purification of D-lactate oxidase using HisTrap Ni affinity chromatography column
[0048] Concentrate the above-mentioned eluate containin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com