Application of novel cyclic peptide
A cyclic peptide and amino acid technology, applied in the field of diseases, can solve the problem that antimicrobial peptides are easily hydrolyzed by proteases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
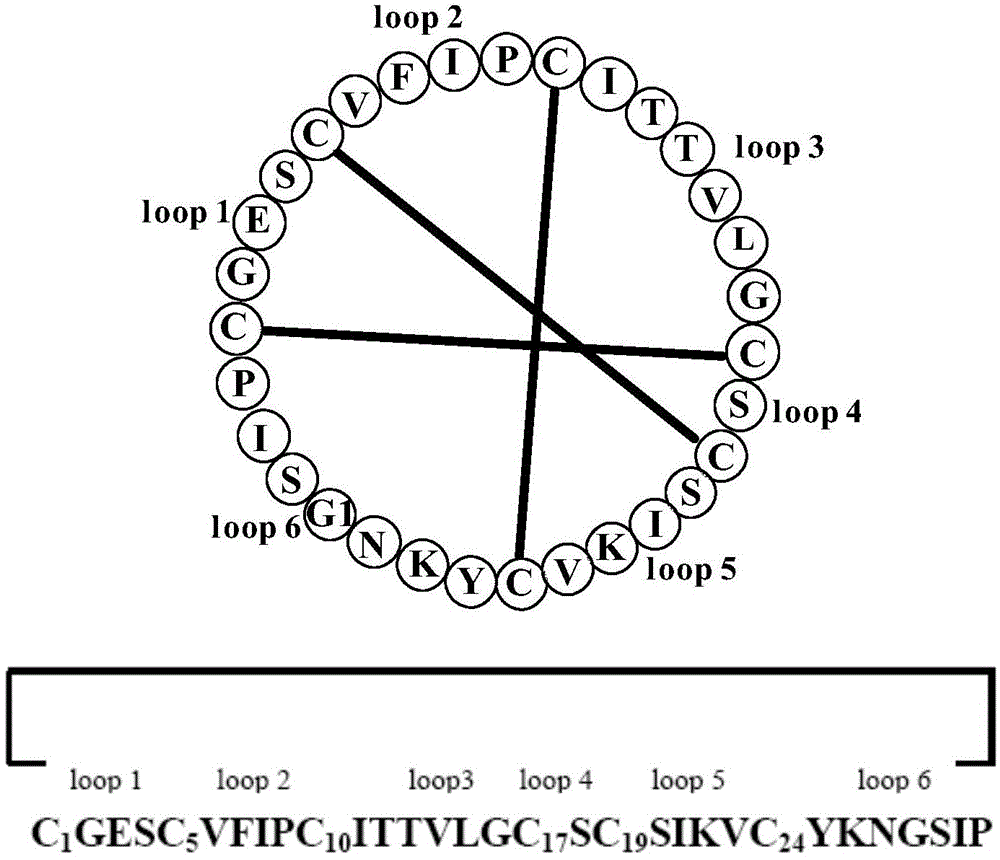
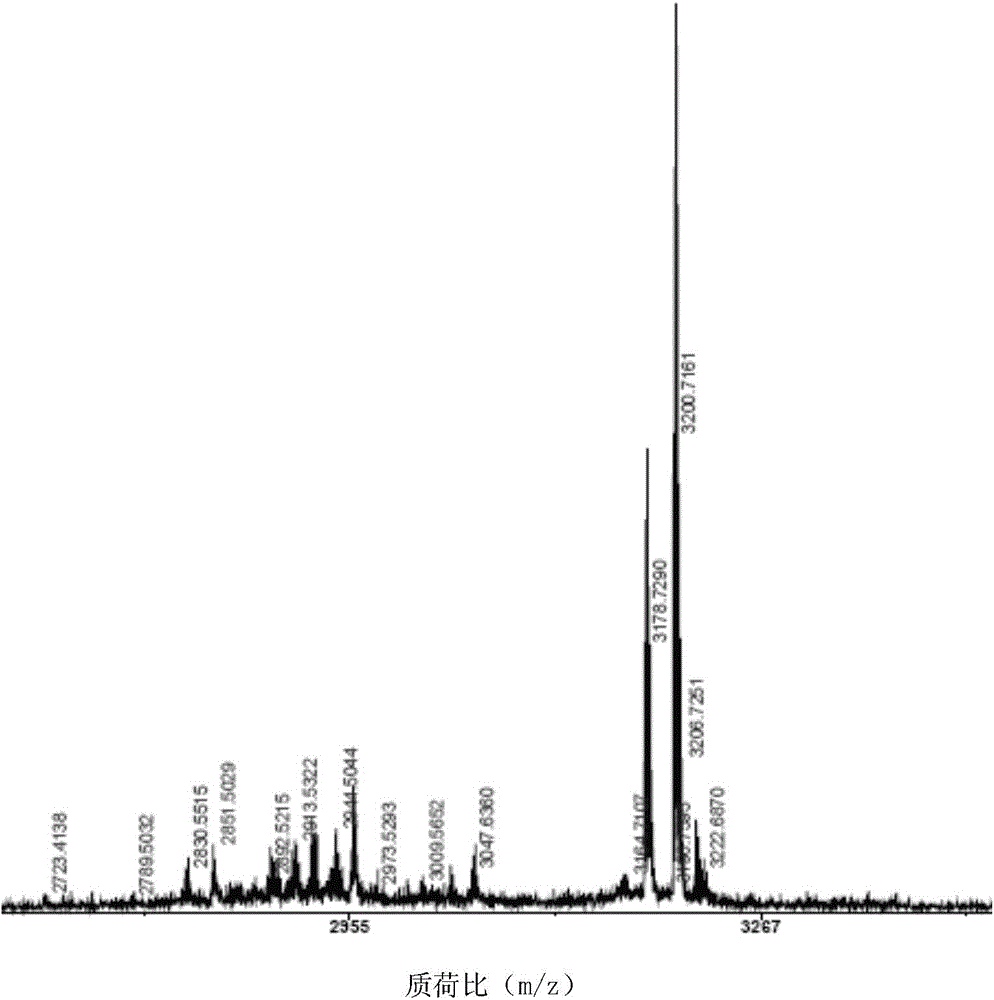
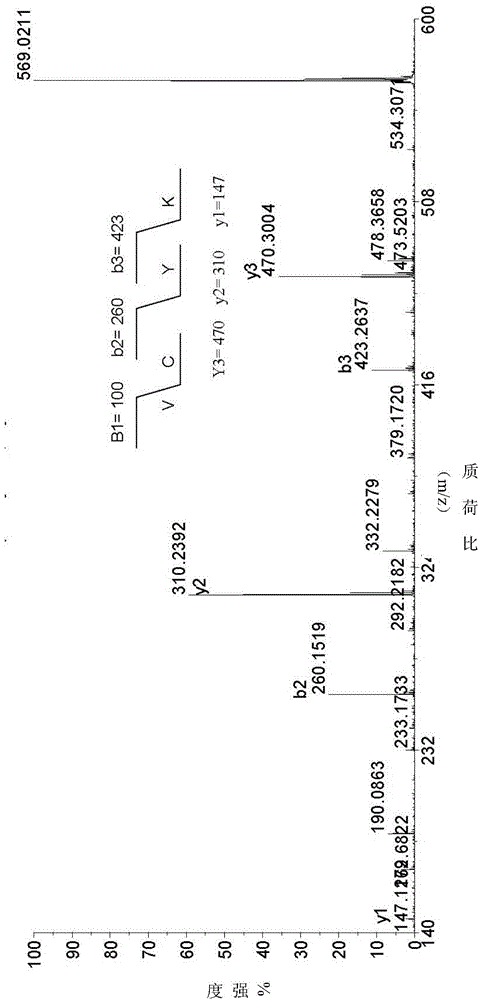
[0065] Separation, Purification and Structure Identification of Natural Cyclic Peptides
[0066] Extraction of samples and separation of cyclic peptides Collect fresh Viola viola, crush it with a juice extractor, then extract it with 95% ethanol for 3 times, each time for 24 hours, combine the extracts, concentrate under reduced pressure and recover ethanol; the extracts are weighed with water Suspended and extracted with petroleum ether, ethyl acetate, and n-butanol in turn; the n-butanol part containing the cyclic peptide was concentrated under reduced pressure and then resuspended in water. The sample was subjected to Sephadex LH-20 column chromatography using 10%, 20%, 30% %, 40%, 50%, 60%, 80% methanol water gradient elution. The eluting fraction containing the cyclic peptide was determined to be the 30% fraction using mass spectrometry tracking. After concentrating again under reduced pressure, use Agilent1200 system and GRACE VYDAC protein polypeptide semi-preparative ...
Embodiment 2
[0077] SEQ ID NO:1 thermal stability test
[0078] (1). Peptide SEQ ID NO: 1 solution: 45 μL of 1 μg / μL polypeptide plus 180 μL of sterile water passed through the membrane, and mix well;
[0079] (2).0min heat resistance reaction test sample: take out 50μL of the mixed solution in step (1);
[0080] (3). Test samples with different heat-resistant reaction times: divide the mixture in step (1) into three samples of 50 μL, place them in a boiling water bath at 100°C, and take out a sample for about 10 minutes, 20 minutes, and 30 minutes respectively. , immediately placed on ice to cool for 10 minutes, and then centrifuged;
[0081] (4). The samples (2) and (3) were analyzed by HPLC using a chromatographic column (YMC-Pack, ODS-A, 250×4.6mm) ( Figure 9 ), the liquid chromatography elution conditions are as follows:
[0082]
[0083] Note: Phase A: 0.5%TFA Phase B: 90%AcN, 0.5%TFA; detection wavelength: 220nm
[0084] (5). Results: With the peak area of 0min SEQ ID NO:1 p...
Embodiment 3
[0087] SEQ ID NO:1 enzymatic stability test
[0088] 1. Solution preparation
[0089] (1). Peptide SEQ ID NO: 1: 1 μg / μL, prepared with sterile water through the membrane;
[0090] (2). Buffer for Trypsin test: 100mM Tris-HCl, pH8.0; Trypsin enzyme: 0.5μg / μL;
[0091] (3). Buffer for Chymotrypsin test: 100mM Tris-HCl, 10mM CaCl 2 ,pH7.8; Chymotrypsin enzyme: 0.5μg / μL;
[0092] (4). Enzyme hydrolysis termination solution: 10% TFA, prepared with corresponding reaction buffer.
[0093] 2. Trypsin enzymatic hydrolysis test of SEQ ID NO:1
[0094] (1). Peptide SEQ ID NO:1 solution: 25 μL 1 μg / μL polypeptide plus 95 μL buffer, mix well;
[0095] (2). Trypsin enzymatic hydrolysis reaction working solution: 0.35 μL 0.5 μg / μL peptide plus 3.15 μL buffer, mix well;
[0096] (3). Place solutions (1) and (2) in a 37°C incubator for 5 minutes;
[0097] (4).0min enzymatic hydrolysis test sample: take out 25 μL of the mixture in step (1), and add 3.75 μL of buffer;
[0098] (5). Test...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com