A kind of expression method of biotinylated inorganic pyrophosphatase
A technology of inorganic pyrophosphatase and expression method, which is applied in the field of expression of biotinylated inorganic pyrophosphatase, and can solve the problems of affecting the catalytic activity of enzyme protein, reducing enzyme activity, loss and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] The expression method of embodiment 1 biotinylated inorganic pyrophosphatase
[0039] Include the following steps:
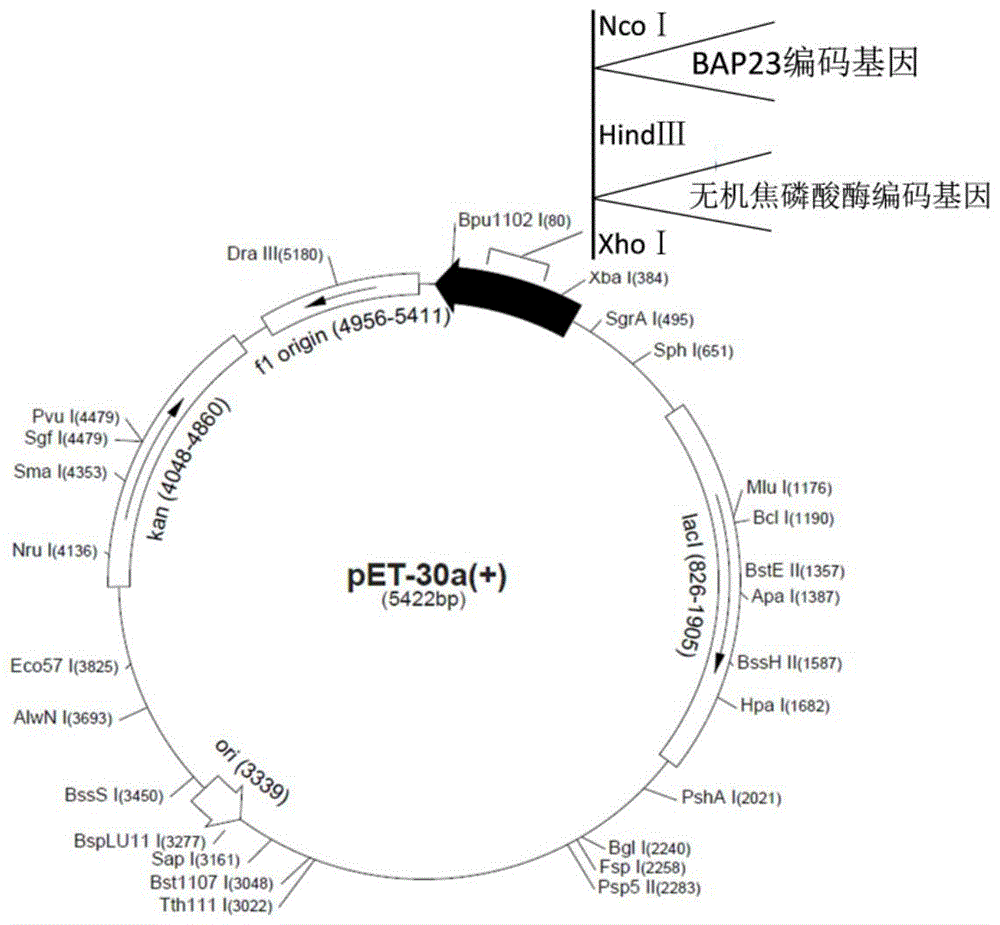
[0040] ⑴. Introducing NcoI and HindIII restriction sites and corresponding protective bases respectively upstream and downstream of the nucleotide sequence shown in SEQ ID NO: 2 to obtain the nucleotide sequence shown in SEQ ID NO: 3, Named the sequence BAP23-1, chemically synthesized sequence BAP23-1;
[0041] (2) Digest the pET30a(+) plasmid and the sequence BAP23-1 obtained in step (1) of Example 1 with restriction endonucleases NcoI and HindIII, and recover the digested product with a DNA gel recovery kit, and digest the nucleotides Sequence BAP23-1 and pET30a(+) plasmids were ligated overnight at 16°C to obtain recombinant expression vector A1;
[0042] ⑶. Design a pair of primer-specific primers, use Thermococcus litoralis genomic DNA as a template, PCR amplify the coding gene of inorganic pyrophosphatase (GenBank number WP_004067338) and recover ...
Embodiment 2
[0046] Embodiment 2 Purification of biotinylated inorganic pyrophosphatase
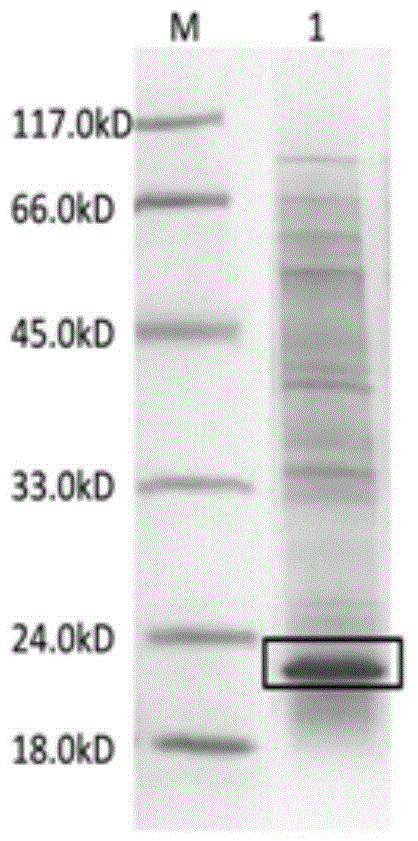
[0047] The biotinylated inorganic pyrophosphatase expressed according to the method of Example 1 was purified by using a histidine affinity chromatography column.
[0048] The purification method comprises the following steps:
[0049] ⑴. Centrifuge to collect the bacterial cells after the induction culture in Step ⑹ of Example 1, crush the bacterial cells with ultrasonic waves, centrifuge the crushed liquid at 4°C and 12000rpm / min for 30min, and collect the supernatant;
[0050] ⑵. Equilibrate the histidine affinity chromatography column with 5 times the volume of the column bed (50mM pH8.0 sodium phosphate buffer + 300mM sodium chloride + 2mM imidazole);
[0051] (3) Add the supernatant obtained in step (1) of Example 2 to a histidine affinity chromatography column at a flow rate of 1 mL / min, so that the biotinylated inorganic pyrophosphatase is attached to the column;
[0052] ⑷. Rinse the chroma...
Embodiment 3
[0055] Embodiment 3 Expression method of biotinylated inorganic pyrophosphatase
[0056] Include the following steps:
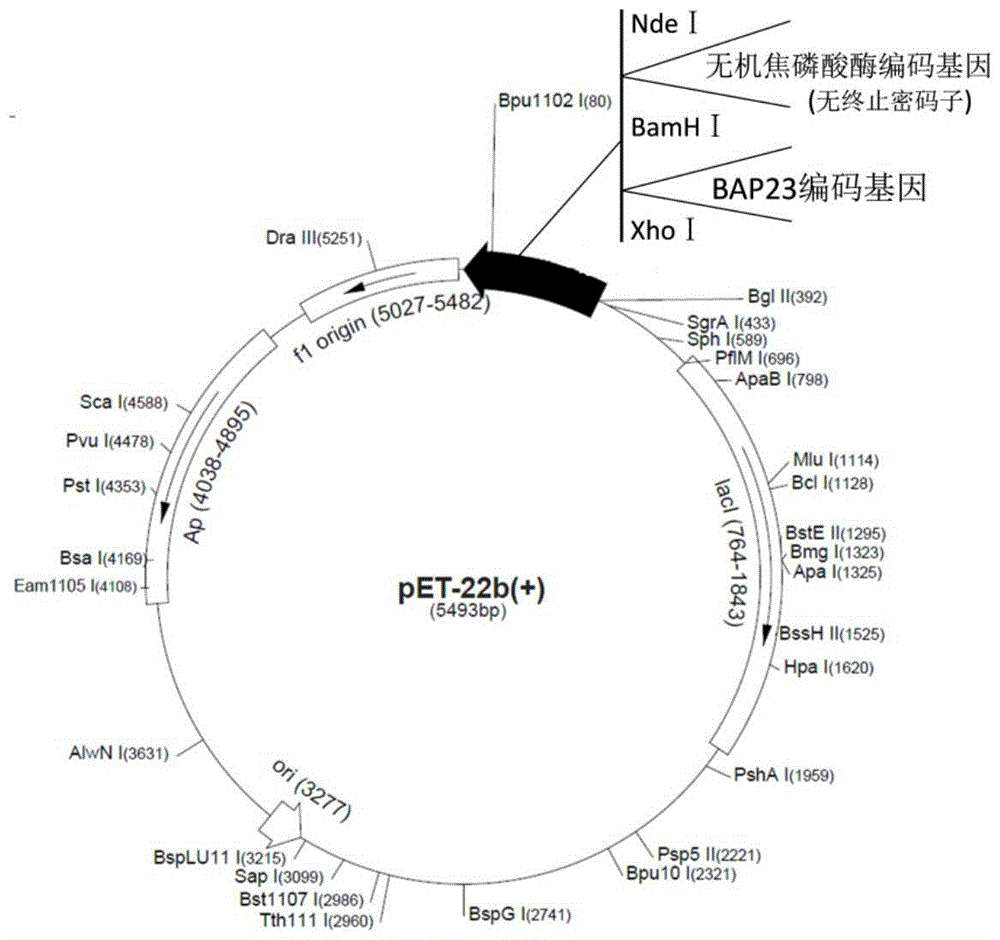
[0057] ⑴. Introducing the restriction sites of BamHI and XhoI and the corresponding protective bases respectively upstream and downstream of the nucleotide sequence shown in SEQ ID NO: 2 to obtain the nucleotide sequence shown in SEQ ID NO: 6, Named as sequence BAP23-2, chemically synthesized sequence BAP23-2;
[0058] (2) Digest the pET22b(+) plasmid and the sequence BAP23-2 obtained in Step 1 of Example 3 with restriction endonucleases BamHI and XhoI, and recover the digested product with a DNA gel recovery kit, and digest the nucleotides Sequence BAP23-2 and pET22b(+) plasmids were ligated overnight at 16°C to obtain recombinant expression vector A2;
[0059] ⑶. Design a pair of primer-specific primers, use Saccharomyces cerevisiae genomic DNA as a template, PCR amplify wherein the coding gene of inorganic pyrophosphatase (GenBank numbering is NP_009565)...
PUM
| Property | Measurement | Unit |
|---|---|---|
| absorbance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com