Kit for identifying bacteria by use of molecular beacon-melting curve technology and application of kit
A molecular beacon probe and kit technology, applied in recombinant DNA technology, microbial assay/test, resistance to vector-borne diseases, etc. Large range, not easy to misjudgment, and the effect of reducing detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Design of Molecular Beacon Probes.
[0044] (1) download the sequence information of 18 kinds of common bacterial 16s-rDNA of clinical infection that can be identified by the kit of the present invention from the database of the National Center for Biotechnology Information (NCBI) in the United States as the basis for the design of molecular beacon probes;
[0045] The 18 common bacteria in clinical infection include the following bacteria: Acinetobacter baumannii, Aeromonas hydrophila, Burkholderia cepacia, Citrobacter freundii, Enterobacter cloacae, Enterococcus faecium, Enterococcus faecalis, Enterobacter, Escherichia coli, Klebsiella pneumoniae, Listeria monocytogenes, Proteus mirabilis, Pseudomonas aeruginosa, Staphylococcus epidermidis, Staphylococcus aureus, Salmonella, Serratia marcescens, Stenotrophomonas maltophilia bacteria.
[0046] (2) Design molecular beacon probes, the principle is: 1. make it cover part of the conserved region sequence and pa...
Embodiment 2
[0058] Example 2: Establishment and optimization of the high-resolution melting curve identification system of the colony identification kit.
[0059] Conditions were optimized for some important influencing factors of the high-resolution melting curve identification system.
[0060] 1. Method
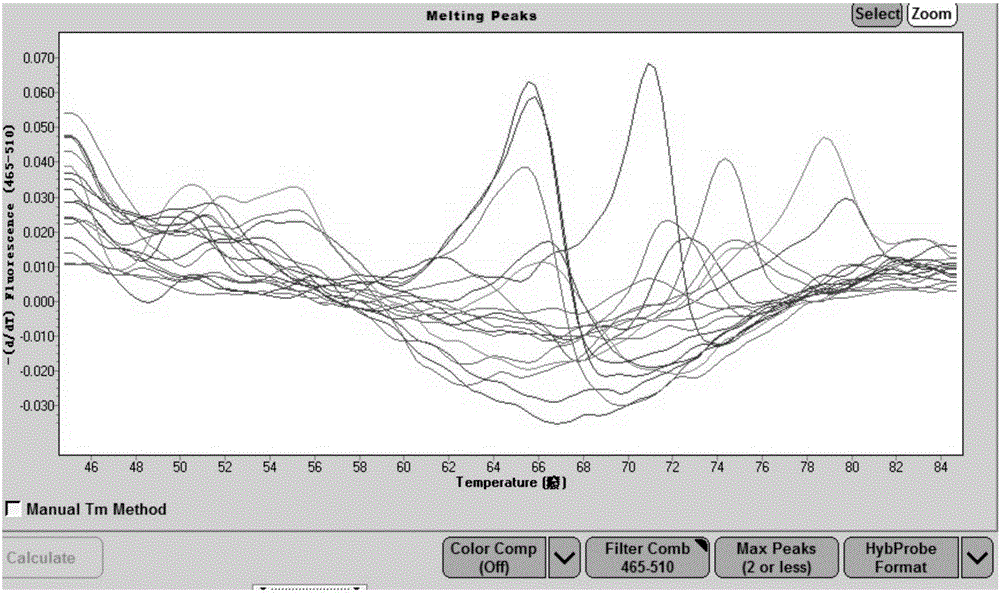
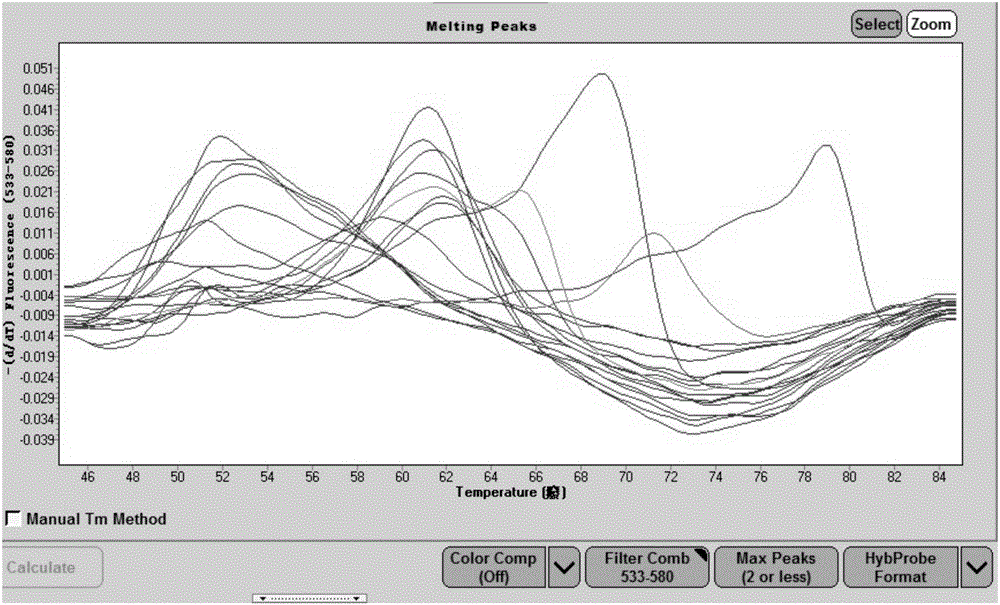
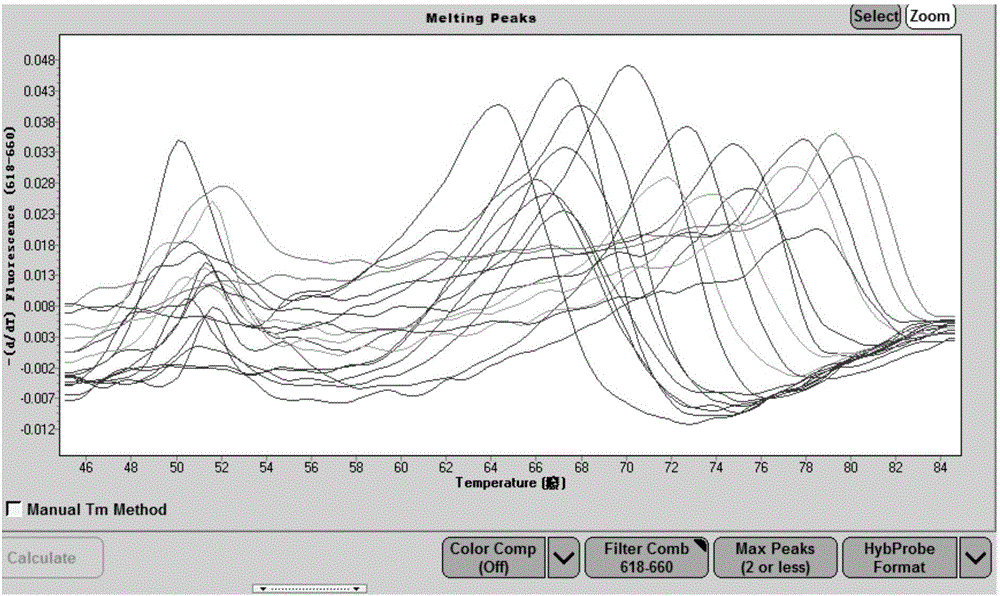
[0061] (1)Mg 2+ Concentration study: Prepare 2x high-resolution melting curve bacterial identification reaction solution according to the reaction system in Table 1, fix other parameters, and adjust Mg respectively 2+ Concentrations up to 0.1M, 0.05M, 0.03M, 0.01M, 0M, high-resolution melting curve analysis using nucleic acid extracted from E. coli standard strains as a template, and comparing different Mg 2+ The effect of concentration on the height, sharpness and fluency of the peak of the melting curve.
[0062] (2)K + Concentration study: Prepare 2× high-resolution melting curve bacterial identification reaction solution according to the reaction system in Table 1, fix other pa...
Embodiment 3
[0069] Embodiment 3: Composition and detection method of high-resolution melting curve bacterial identification kit
[0070] 1. Composition of the kit (stored at -20°C)
[0071] (1) 2× high resolution melting curve reaction solution: its components are: 0.4M betaine, 1.5% (V / V) DMSO, 80mM
[0072] Tris-HCl, 200 mM potassium chloride, 80 mM magnesium chloride.
[0073] 2. Preparation of positive control substance
[0074] Escherichia coli strain CICC10421=078:K80 was inoculated on a Columbia blood plate, cultured at 37° C. and 70% RH overnight, and nucleic acid was extracted from colonies. Measure the concentration and purity of the extracted nucleic acid to ensure that 1.8 / 1.6 is not lower than 1.8; then adjust the nucleic acid concentration to 50ng / ul with inactivated pure water. Store at -20°C after aliquoting as a positive control for the kit.
[0075] 3. The detection method of the kit is as follows:
[0076] (1) extract the nucleic acid of the sample, which contains ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com