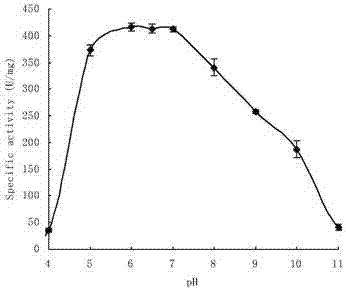
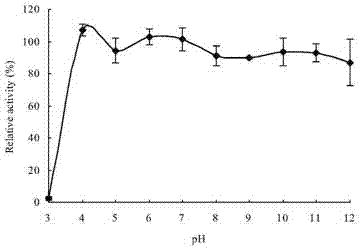
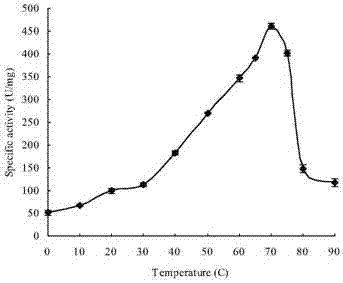
Neutral high temperature xylanase as well as coding genes and application thereof
A technology encoding gene and xylanase, applied in the field of genetic engineering, can solve the problems of high energy and acid-base chemical reagents, environmental pollution, cost increase, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Cloning of the genomic DNA of embodiment 1, neutral high-temperature xylanase CtXyn10A
[0074] Extract the genomic DNA of Cladosporium tianshanense SL-14:
[0075] Filter the mycelium cultured in liquid for 3 days with sterile filter paper, put it into a mortar, add 2mL of extract, grind for 5min, then put the grinding solution in a 50mL centrifuge tube, lyse in a water bath at 65°C for 20min, and mix every 10min. Homogenize once and centrifuge at 10,000 rpm for 5 min at 4°C. The supernatant was extracted in phenol / chloroform to remove impurity proteins, and then an equal volume of isopropanol was added to the supernatant. After standing at room temperature for 5 minutes, centrifuge at 10,000 rpm for 10 minutes at 4°C. The supernatant was discarded, the precipitate was washed twice with 70% ethanol, dried in vacuum, dissolved by adding an appropriate amount of TE, and stored at -20°C for later use.
[0076] According to the sequence analysis of the genome framework o...
Embodiment 2
[0083] Example 2, Acquisition of the gene encoding mature neutral high-temperature xylanase CtXyn10A
[0084] Extract the total RNA of Cladosporium tianshanense SL-14, use reverse transcriptase to obtain a strand of cDNA, and then use primers Ctxyn10A-F / Ctxyn10A-R to amplify the single-stranded cDNA, and the amplified product is recovered and sent to III Sequenced by Bo Biotechnology Co., Ltd.
[0085] Ctxyn10A-F: 5'-GGGGAATTCCACCCTTCGGCTCCCAAGGAC-3';
[0086] Ctxyn10A-R: 5'-GGGGCGGCCGCTCACTTCTTGCCACCCTTGATGCC-3';
[0087] After comparing the genome sequence of the nucleic acid fragment measured in Example 1 with the sequencing result of the above-mentioned PCR amplification product, it was found that the gene has 2 introns, the cDNA is 1059bp long, and has the nucleotide of SEQ ID No. 2 in the sequence table Sequence, the amino acid sequence shown in SEQ ID No. 3 in the coding sequence list and a stop codon. A protein with the amino acid sequence shown in SEQ ID №: 3 in th...
Embodiment 3
[0090] Embodiment 3, the preparation of neutral high-temperature xylanase CtXyn10A
[0091] The Pichia pastoris expression vector pPIC9 was subjected to double digestion (EcoRI+NotI), and the nucleic acid fragment encoding the mature protein prepared in Example 2 was double-digested (EcoRI+NotI) to cut out the gene fragment encoding the mature protein and The expression vector pPIC9 after double digestion was ligated.
[0092] Sequencing to verify the correctness of the sequence. The sequence of the foreign gene inserted in the resulting recombinant plasmid is SEQ ID No.: 2 nucleotides 55-1059, and the recombinant plasmid is named pPIC-Ctxyn10A.
[0093] The above-mentioned recombinant plasmid pPIC-Ctxyn10A was transformed into Pichia pastoris GS115; the correctness of the sequence was verified by extracting the plasmid from the positive recombinant bacteria, and the recombinant Pichia strain containing the recombinant plasmid pPIC-Ctxyn10A was named GS115 / Ctxyn10A.
[0094]...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com