Nucleic acid reagent, kit and system for detecting lower respiratory tract infection bacteria
A respiratory and kit technology, applied in the direction of microorganism-based methods, microorganism measurement/inspection, biochemical equipment and methods, etc., can solve problems such as poor ability to produce antibodies, inability to meet fast detection, and inapplicability of early diagnosis, etc. to save time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Embodiment 1 detection method and detection result judgment
[0073] 1. Primer and probe synthesis
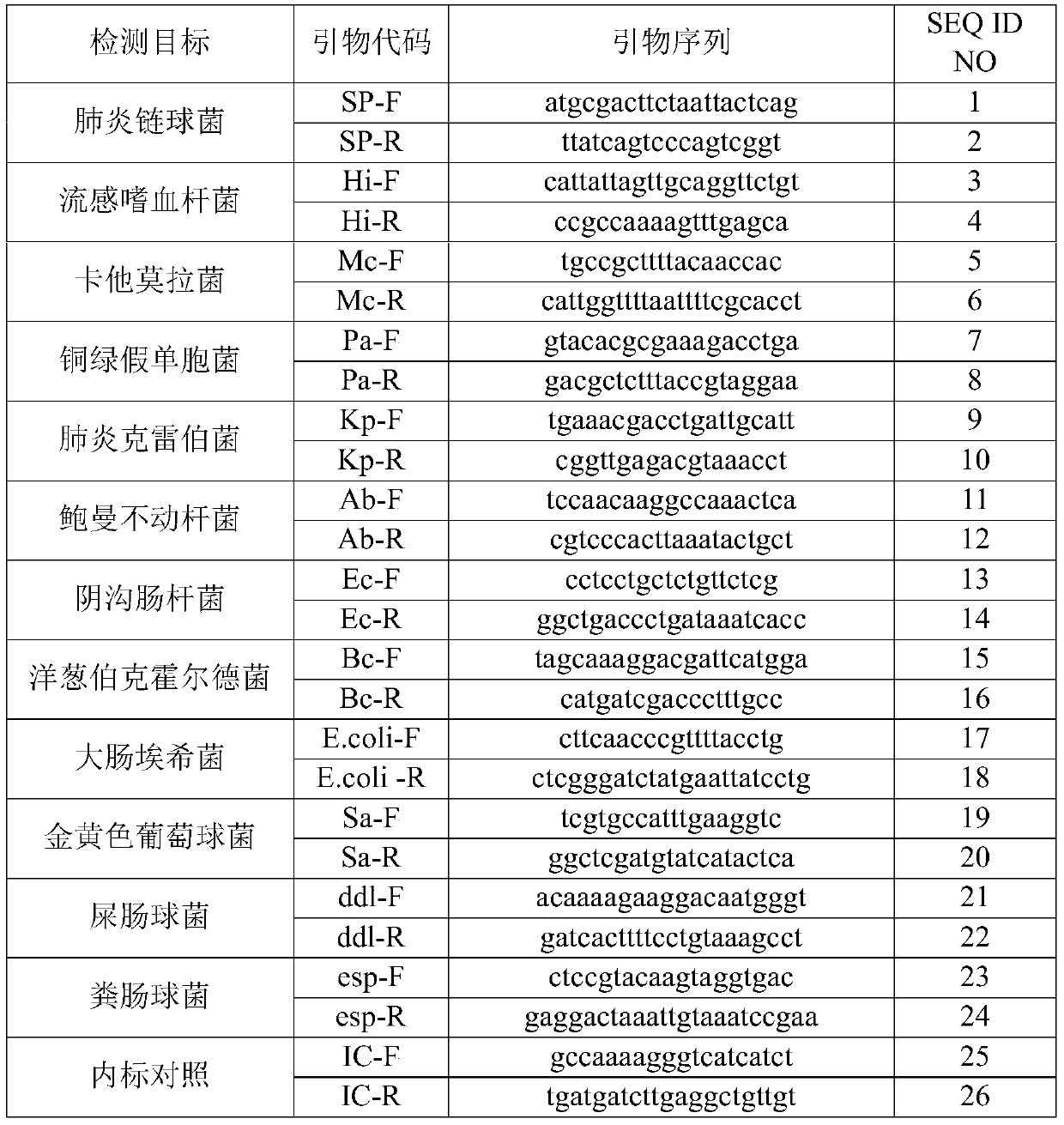
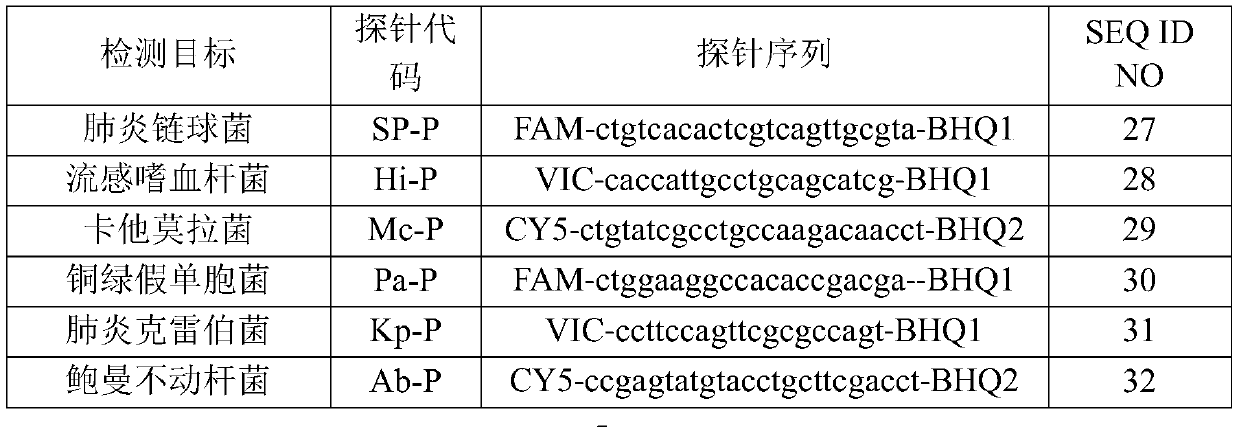
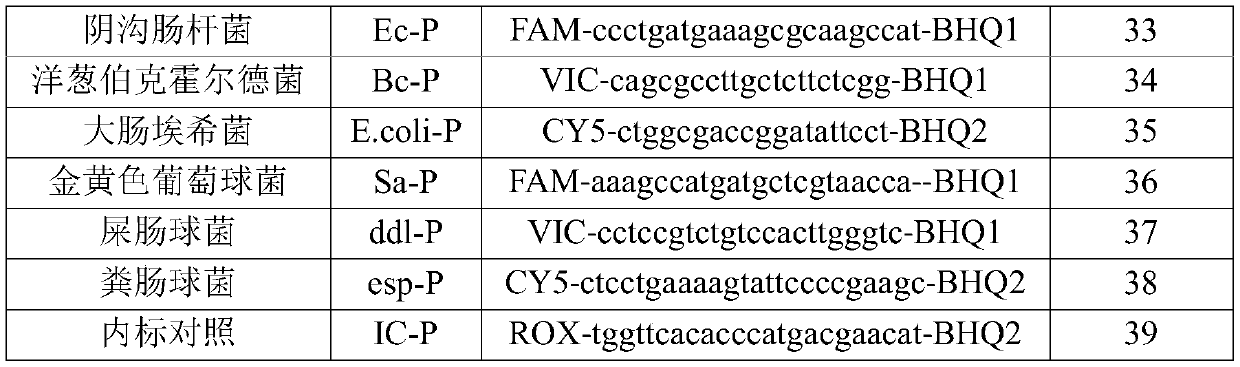
[0074] Sequence synthesis was performed according to the primer sequences shown in Table 1 and the probe sequences shown in Table 2. In the probe, FAM is 6-carboxyfluorescein, CY5 is 5H-indocyanine, ROX is 6-carboxy-X-rhodamine, VIC is a dye purchased from ABI, and BHQ1 and BHQ2 are quenching groups.
[0075] Table 1
[0076]
[0077] Table 2
[0078]
[0079]
[0080] 2. Sample processing
[0081] The liquefied sputum sample was centrifuged at 13,000 rpm for 5 minutes, and the centrifuged precipitate was mixed with 200 μL of normal saline, then centrifuged and washed at 13,000 rpm for 5 minutes, and repeated twice. After centrifugation and washing, remove the supernatant, add 50 μL nucleic acid extraction solution and 10 μL internal standard control to the obtained precipitate, shake and mix, heat in a metal bath or boiling water bath at 100 °C for 10 minut...
Embodiment 2
[0099] Embodiment 2 minimum detection limit verification
[0100] Test samples for evaluation: Use commercial kits to extract Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Enterobacter cloacae, Genomic DNA from Burkholderia cepacia, Escherichia coli, Staphylococcus aureus, Enterococcus faecium, and Enterococcus faecalis, and quantified at 1 × 10 6 copies / mL, and then serially diluted to obtain a concentration of 1×10 5 copies / mL, 1×10 4 copies / mL, 1×10 3 copies / mL, 1×10 2 Copies / mL evaluation template.
[0101] According to the detection method of Example 1, the evaluation templates of each concentration were detected respectively, and the detection was repeated 20 times for each concentration gradient, and the average value was taken as the final detection result, as shown in Table 3.
[0102] table 3
[0103]
[0104] Note: "+" means positive, n / 20 means the detection ...
Embodiment 3
[0106] Example 3 specificity verification
[0107]Select human coronavirus (purchased from Wuhan Institute of Virology, Chinese Academy of Sciences, number 2233), cytomegalovirus (purchased from Wuhan Institute of Virology, Chinese Academy of Sciences, number 303), enterovirus (purchased from Wuhan Institute of Virology, Chinese Academy of Sciences, number 977) ), measles virus (purchased from Wuhan Institute of Virology, Chinese Academy of Sciences, number 1513), human metapneumovirus (from the General Hospital of the Chinese People's Liberation Army, number 1768), rhinovirus (purchased from Wuhan Institute of Virology, Chinese Academy of Sciences, number 2343 ), avirulent Mycobacterium tuberculosis (purchased from China Medical Culture Collection Center, No. 93009), Neisseria meningitidis (from the General Hospital of the Chinese People's Liberation Army, No. 21123), Streptococcus pyogenes (from PLA General Hospital, No. 25341), Streptococcus salivarius (from the Chinese Peo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com