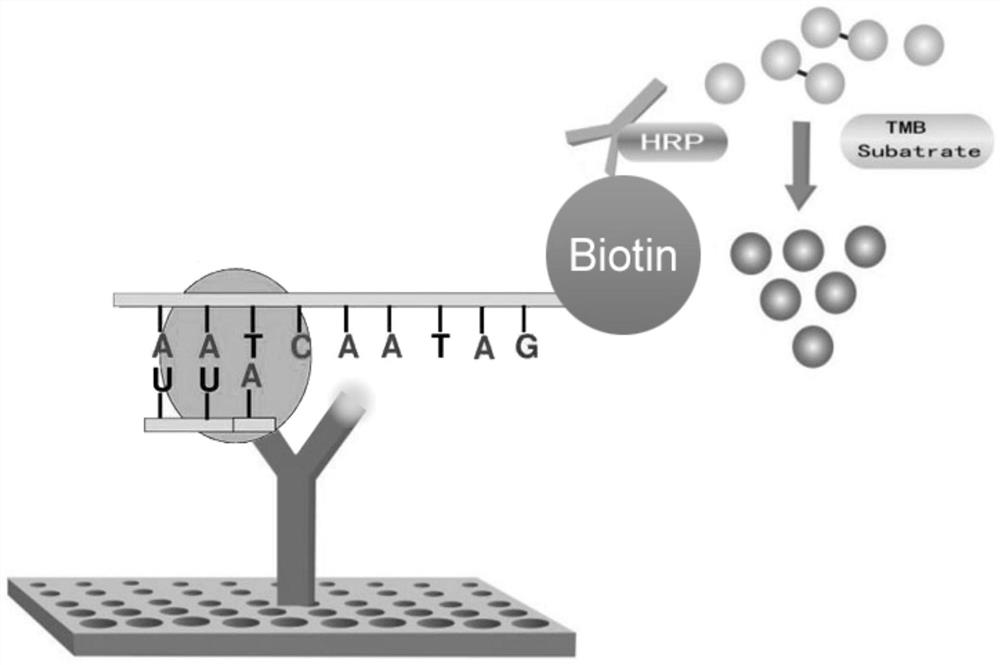
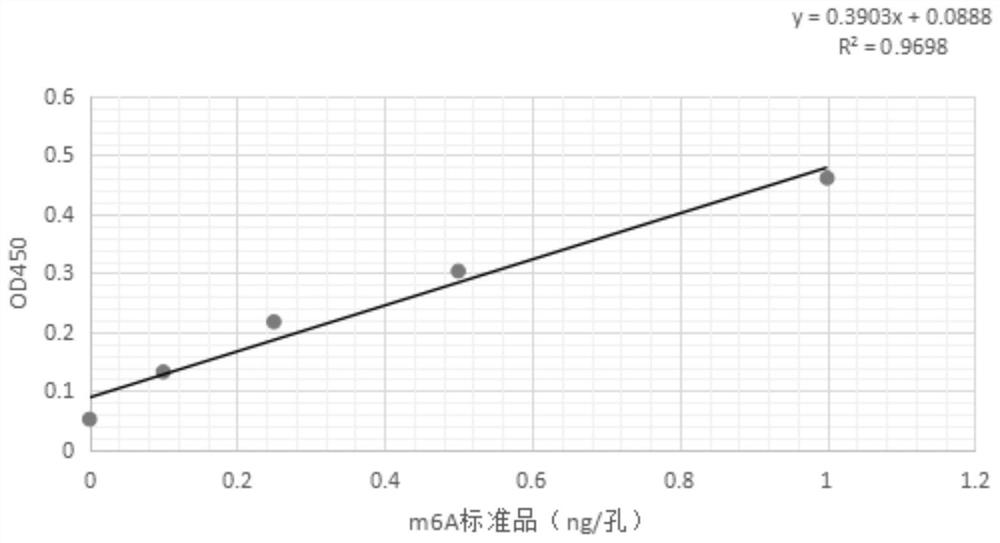
RNA methylation m6A detection method and reagent kit
A detection kit and detection method technology, applied in the field of molecular biology, can solve the problems of inability to quantify modified sites and inaccurate positioning of adenine nucleotides, etc., achieve repeatability and reliability, save experimental funds, and be easy to operate Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
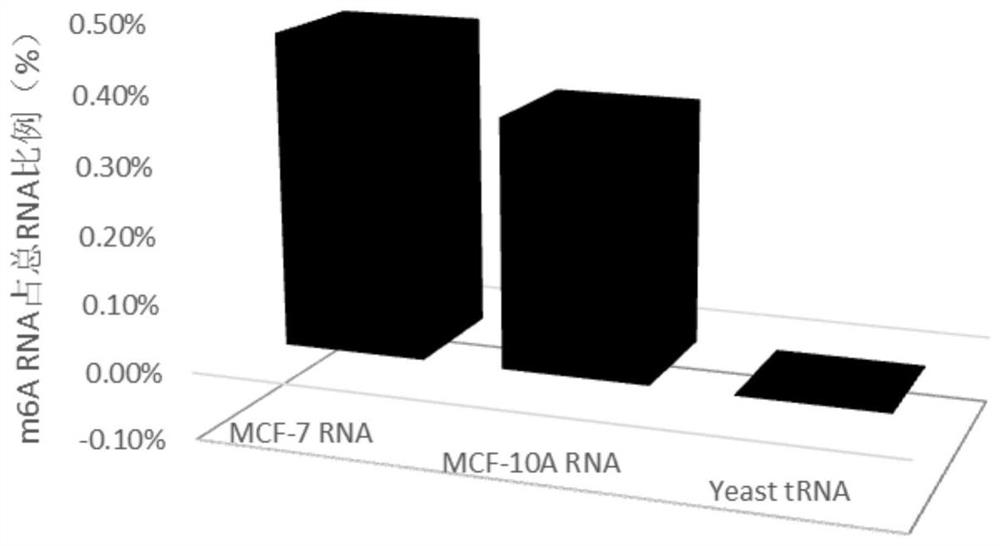
[0048] Extract human breast cancer cell RNA from serum, take 200ng of human breast cancer cell RNA, and take the following steps for processing:
[0049] 1 Sample collection and preparation
[0050] 1.1 Take human breast cancer cells, lyse and extract RNA.
[0051] 1.2 Purity test: 1 μl RNA sample was diluted 50 times, and the OD value was measured on the BioPhotometer plus Eppendorf Nucleic Acid Protein Analyzer. The ratio of OD260 / OD280 was greater than 1.8, indicating that the prepared RNA was relatively pure and free of protein contamination;
[0052]1.3 Integrity detection of total RNA: take 1 μl of RNA sample, run 1% agarose gel electrophoresis at 80V×20min, observe the 5s rRNA, 18s rRNA and 28s rRNA bands of the total RNA with a gel imaging system, if the three bands are complete It proved that the extraction of total RNA was relatively complete.
[0053] 2. Experimental pretreatment steps
[0054] 2.1 Before the test, place the reagent and the sample to be tested at...
Embodiment 2
[0083] Grind the brain tissue and extract RNA, take 200ng of the obtained brain cell RNA, and take the following steps for processing:
[0084] 1 Sample collection and preparation
[0085] 1.1 Take human brain tissue cells, lyse and extract RNA.
[0086] 1.2 Purity test: 1 μl RNA sample was diluted 50 times, and the OD value was measured on the BioPhotometer plus Eppendorf Nucleic Acid Protein Analyzer. The ratio of OD260 / OD280 was greater than 1.8, indicating that the prepared RNA was relatively pure and free of protein contamination;
[0087] 1.3 Integrity detection of total RNA: take 1 μl of RNA sample, run 1% agarose gel electrophoresis at 80V×20min, observe the 5s rRNA, 18s rRNA and 28s rRNA bands of the total RNA with a gel imaging system, if the three bands are complete It proved that the extraction of total RNA was relatively complete.
[0088] 2. Experimental pretreatment steps
[0089] Operate with reference to the steps in Example 1.
[0090] 3 Sample addition a...
Embodiment 3
[0093] Take 200ng of yeast tRNA and take the following steps for processing:
[0094] 1 Sample collection and preparation
[0095] 1.1 Purity test: 1 μl RNA sample was diluted 50 times, and the OD value was measured on the BioPhotometer plus Eppendorf Nucleic Acid Protein Analyzer. The ratio of OD260 / OD280 was greater than 1.8, indicating that the prepared RNA was relatively pure and free of protein contamination;
[0096] 1.2 Integrity detection of total RNA: Take 1 μl of RNA sample, run 1% agarose gel electrophoresis at 80V×20min, observe the 5s rRNA, 18s rRNA and 28s rRNA bands of the total RNA with a gel imaging system, if the three bands are complete Prove that the total RNA extraction is relatively complete.
[0097] 2. Experimental pretreatment steps
[0098] Operate with reference to the steps in Example 1.
[0099] 3 Sample addition and incubation steps
[0100] Operate with reference to the steps in Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com