Method for degrading protein by co-culture of proteiniphilum acetatigenes and geobacter sulfurreducens
A technology of acetic acidophilic protein and sulfur reduction site, applied in the field of microorganisms, can solve the problems of complicated treatment, high cost, waste of nutrients, etc., and achieve the effect of simple and convenient operation, short treatment time, and promotion of degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment A1
[0032] Take embodiment A1 as an example:
[0033] S1: Co-cultivation of two bacteria:
[0034] A: Preparation of culture medium: first add 500mL distilled water to a 1L Erlenmeyer flask, then add poly-peptone 2.5g / L, yeast extract 5.0g / L, tryptone 2.5g / L, glucose 10.0g / L, chloride Ammonium 1.5g / L, calcium chloride 0.024g / L, potassium chloride 0.1g / L, sodium dihydrogen phosphate 0.64g / L, dipotassium hydrogen phosphate 0.04g / L, vitamin solution 5mL, trace element solution 5mL, and then Add pure water to make the volume up to 1L, heat it in a water bath until it dissolves, add NaOH dropwise to adjust the pH to 6.9, then repack it into narrow-mouth bottles, insert the aeration needle into the narrow-mouth bottle, and inject 80% N 2 and 20%H 2 Mixed gas, aerated for 20 minutes, quickly capped the rubber stopper, placed the narrow-mouth bottle in an autoclave, sterilized at 121°C for 20 minutes, and then transferred to the ultra-clean bench for ultraviolet irradiation;
[0035] B...
Embodiment A1-A4
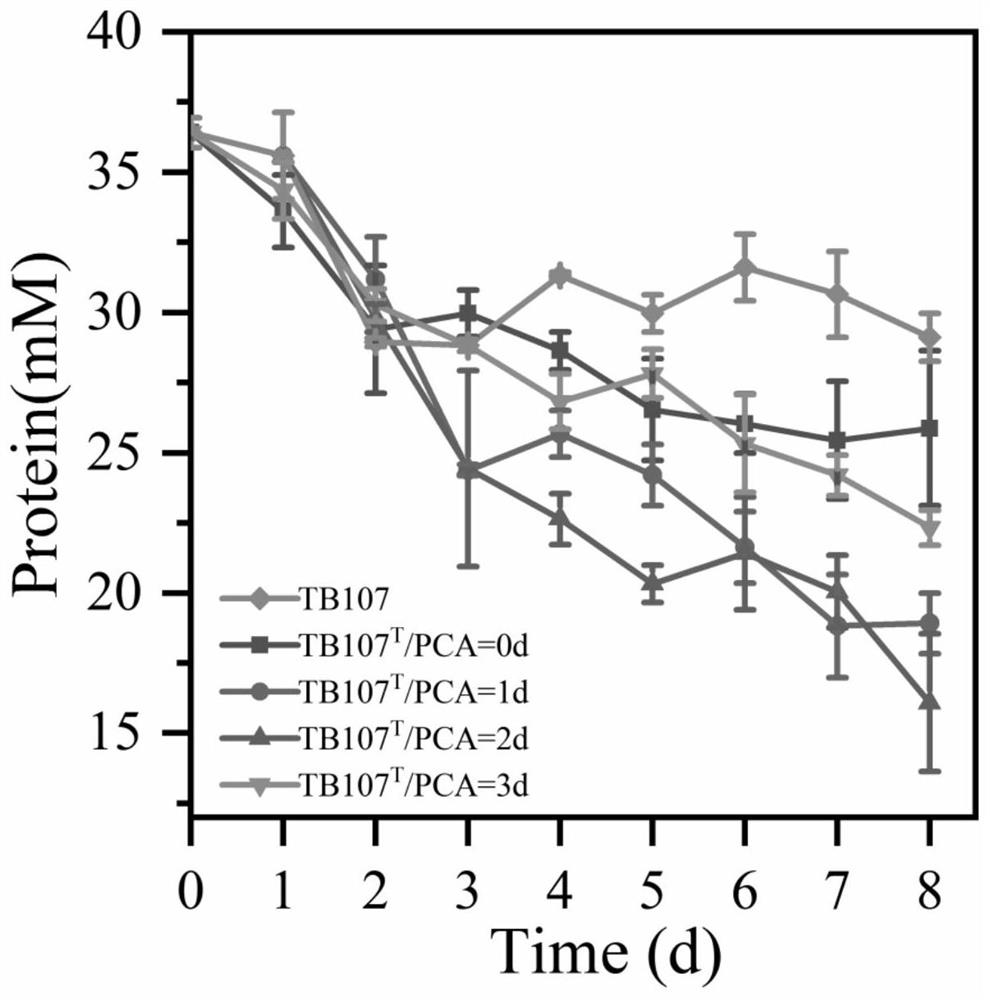
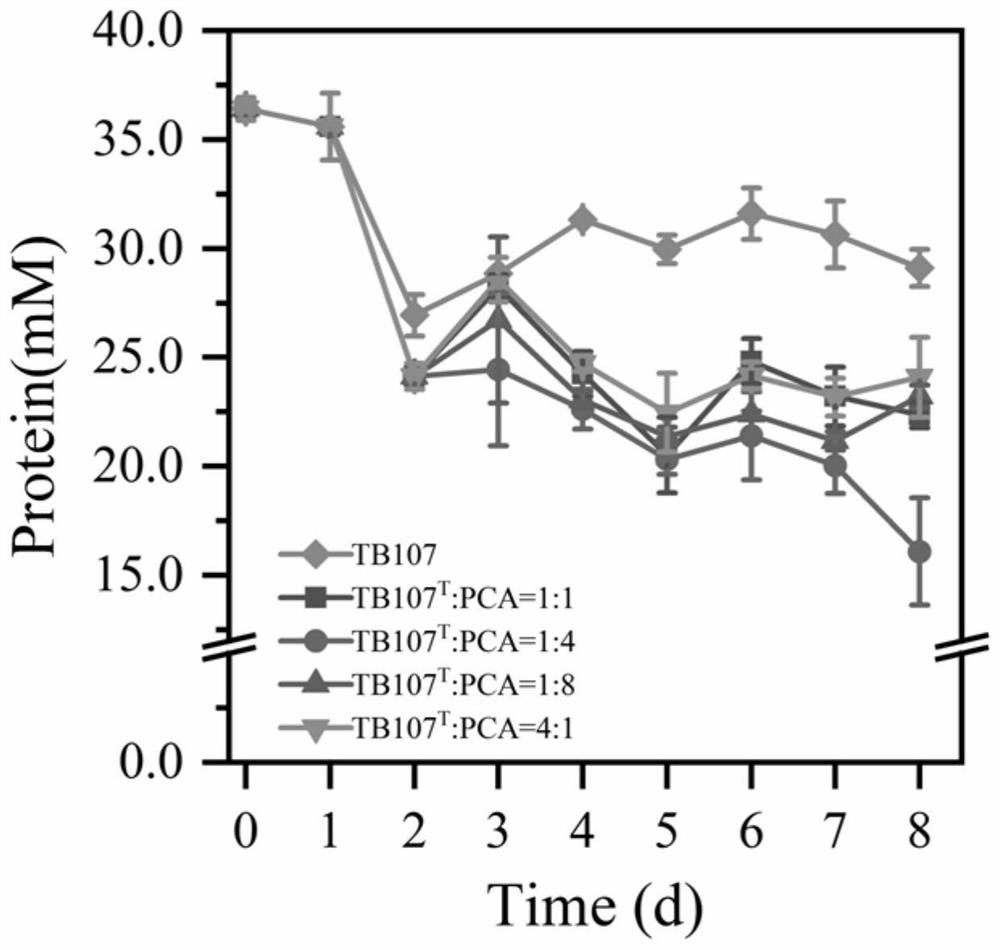
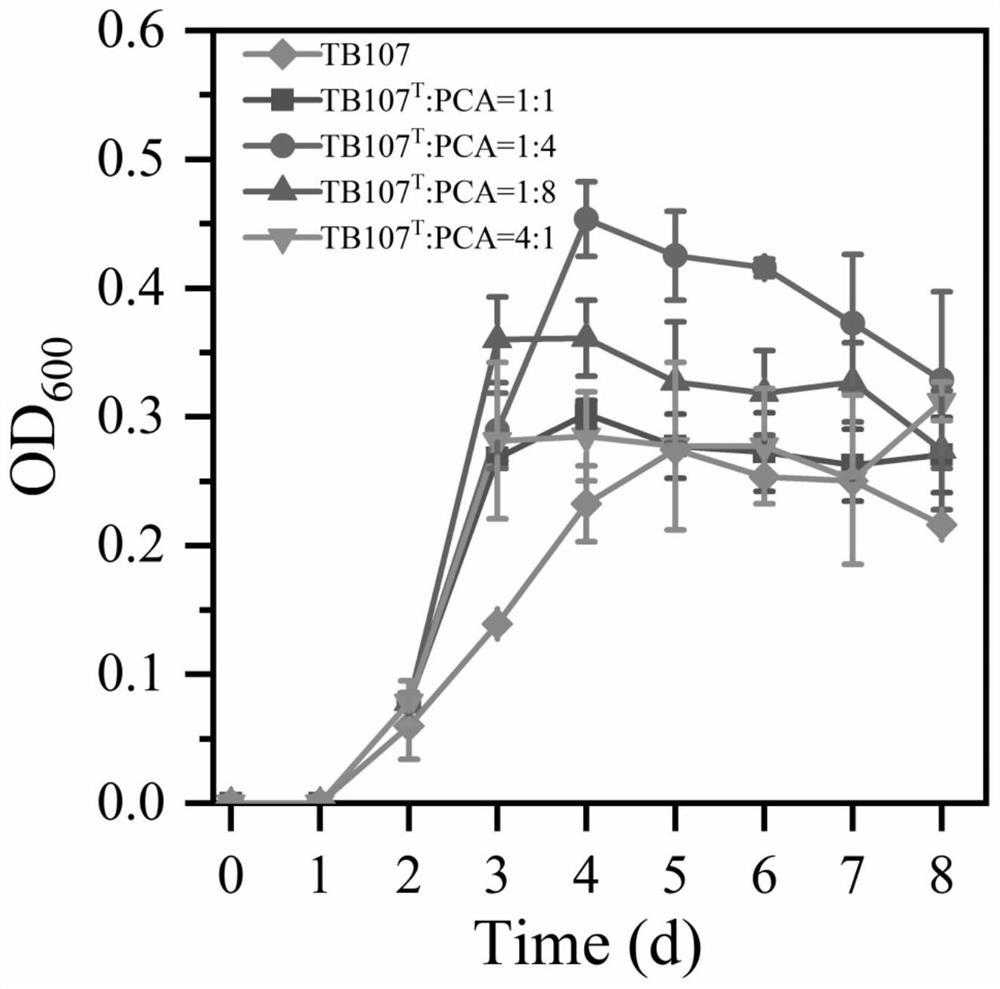
[0040] Embodiments A1-A4: According to the replacement of the inoculation time in the embodiment A1, the specific data are shown in Table 1, and the protein degradation rate was detected on the mixed bacterial solution, and the specific data are shown in Table 2.
[0041] Table 1 embodiment A1-A4 mixed bacterial liquid inoculation time
[0042] A1 A2 A3 A4 Inoculation time 0 1 2 3
Embodiment B1
[0047] Take embodiment B1 as an example:
[0048] S1: Co-cultivation of two bacteria:
[0049] A: Preparation of culture medium: first add 500mL distilled water to a 1L Erlenmeyer flask, then add poly-peptone 2.5g / L, yeast extract 5.0g / L, tryptone 2.5g / L, glucose 10.0g / L, chloride Ammonium 1.5g / L, calcium chloride 0.024g / L, potassium chloride 0.1g / L, sodium dihydrogen phosphate 0.64g / L, dipotassium hydrogen phosphate 0.04g / L, vitamin solution 5mL, trace element solution 5mL, and then Add pure water to make the volume up to 1L, heat it in a water bath until it dissolves, add NaOH dropwise to adjust the pH to 7.0, then repack it into a narrow-mouthed bottle, insert the aeration needle into the narrow-mouthed bottle, and inject 80% N 2 and 20%H 2 Mixed gas, aerated for 20 minutes, quickly capped the rubber stopper, placed the narrow-mouth bottle in an autoclave, sterilized at 121°C for 20 minutes, and then transferred to the ultra-clean bench for ultraviolet irradiation;
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com