Double function kes carrier suitable for streptomycete chromosome gene knock-out
A chromosomal gene and Streptomyces technology, applied in the field of gene carrier, can solve problems such as restriction of exogenous DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Construction of the chromosomal gene knockout structure pHZ1553 of Streptomyces nanchang
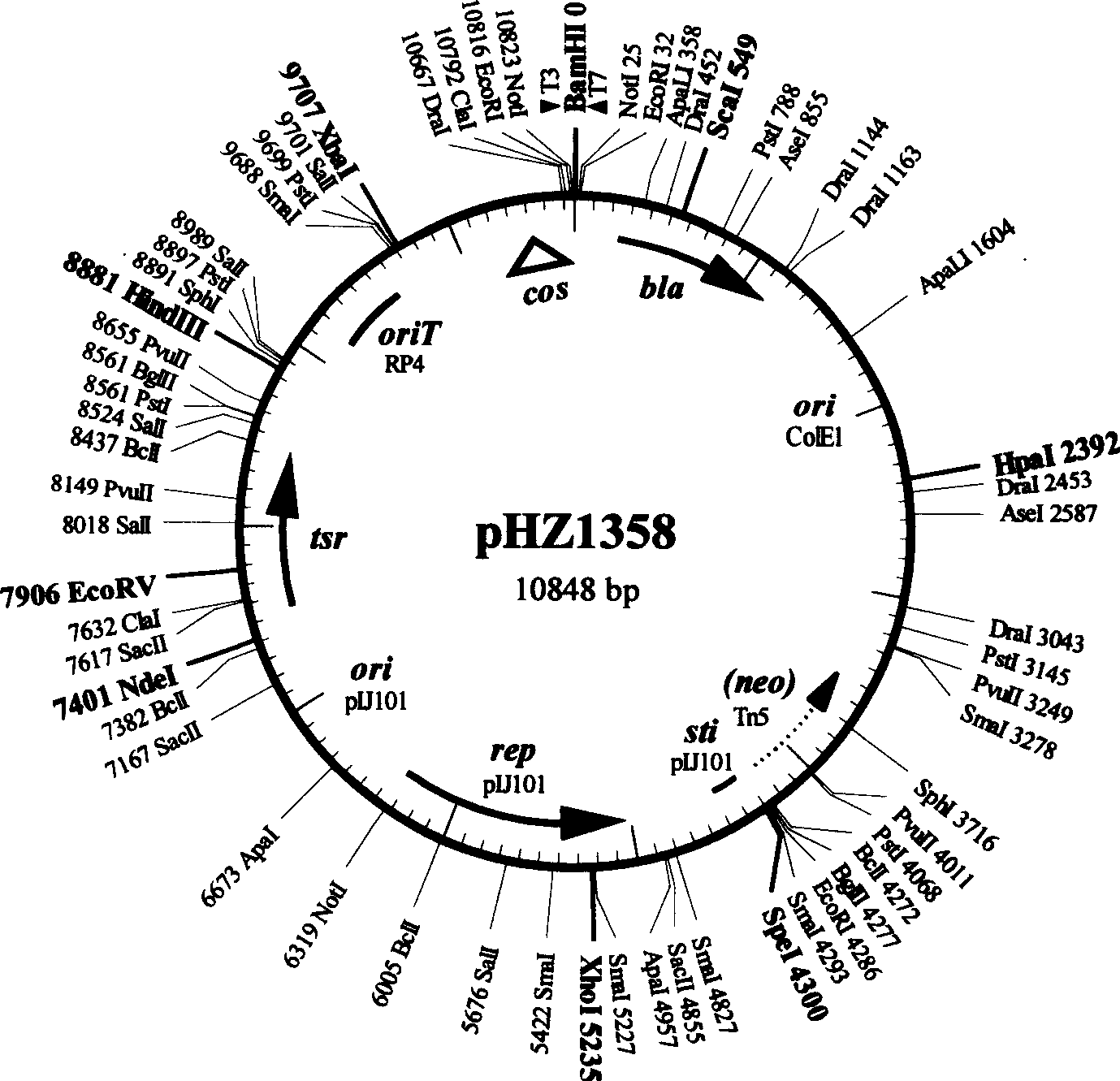
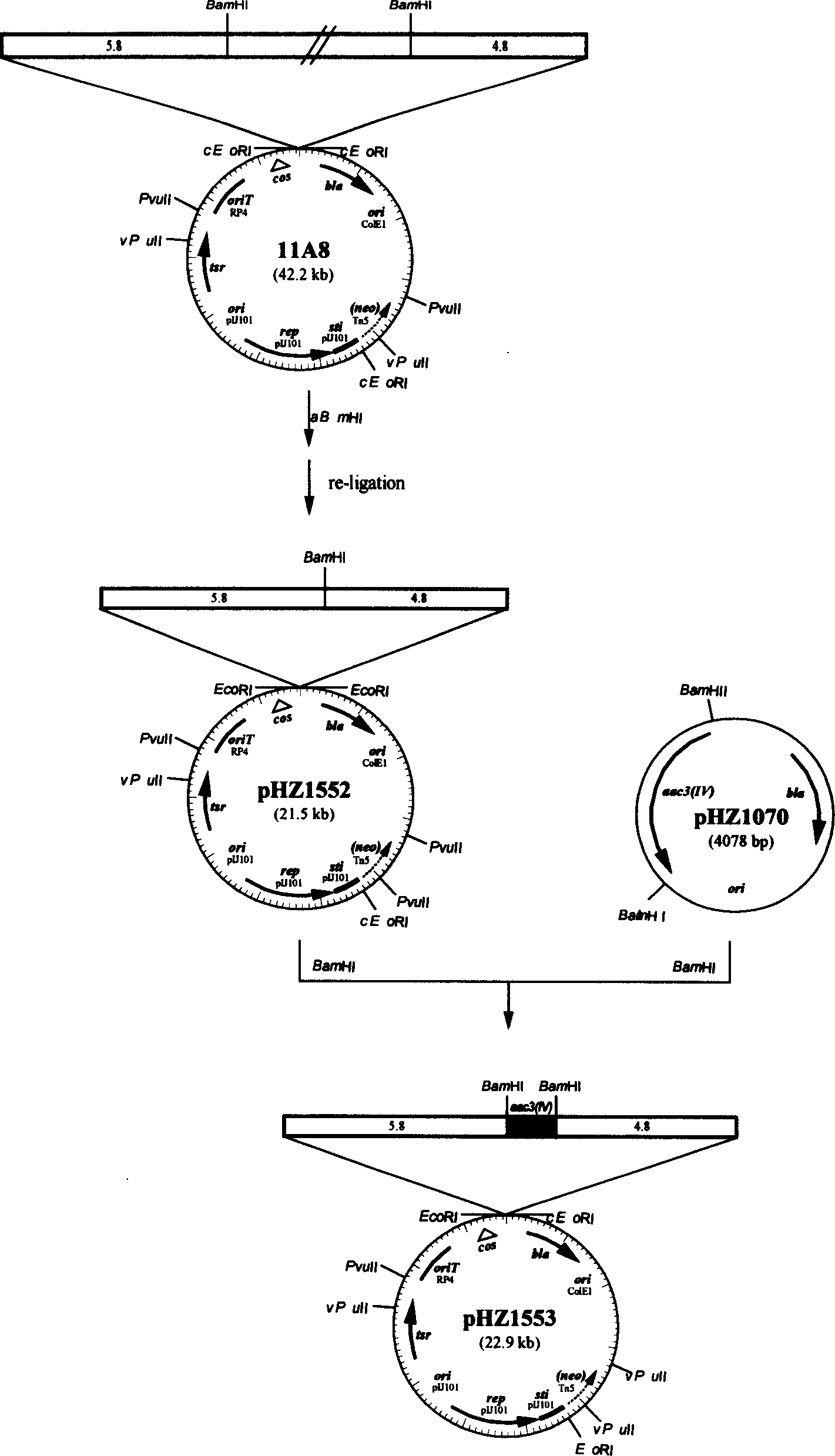
[0033] First, the cosmid 11A8 (in pHZ1358( figure 1 A DNA fragment of about 37 kb from the chromosome of Streptomyces nanchang was inserted into the BamHI single restriction site of ), and this cosmid was completely digested with BamHI, and the digested product was purified and self-ligated at a low concentration, and transformed into DH5α, Quickly check transformants to pick clones with minimal inserts. The size of the inserted fragment was detected by double digestion with BamHI+EcoRI. The sizes of the two foreign fragments were 4.8kb and 5.8kb respectively, and they were named pHZ1552. pHZ1070 was completely digested with BamHI, and the 1.4kb apramycin resistance gene aac3(IV) was recovered and inserted into the BamHI single restriction site of pHZ1552 as a selection marker for gene knockout, thus completing the target gene knockout structure pHZ1553 build. See th...
Embodiment 2
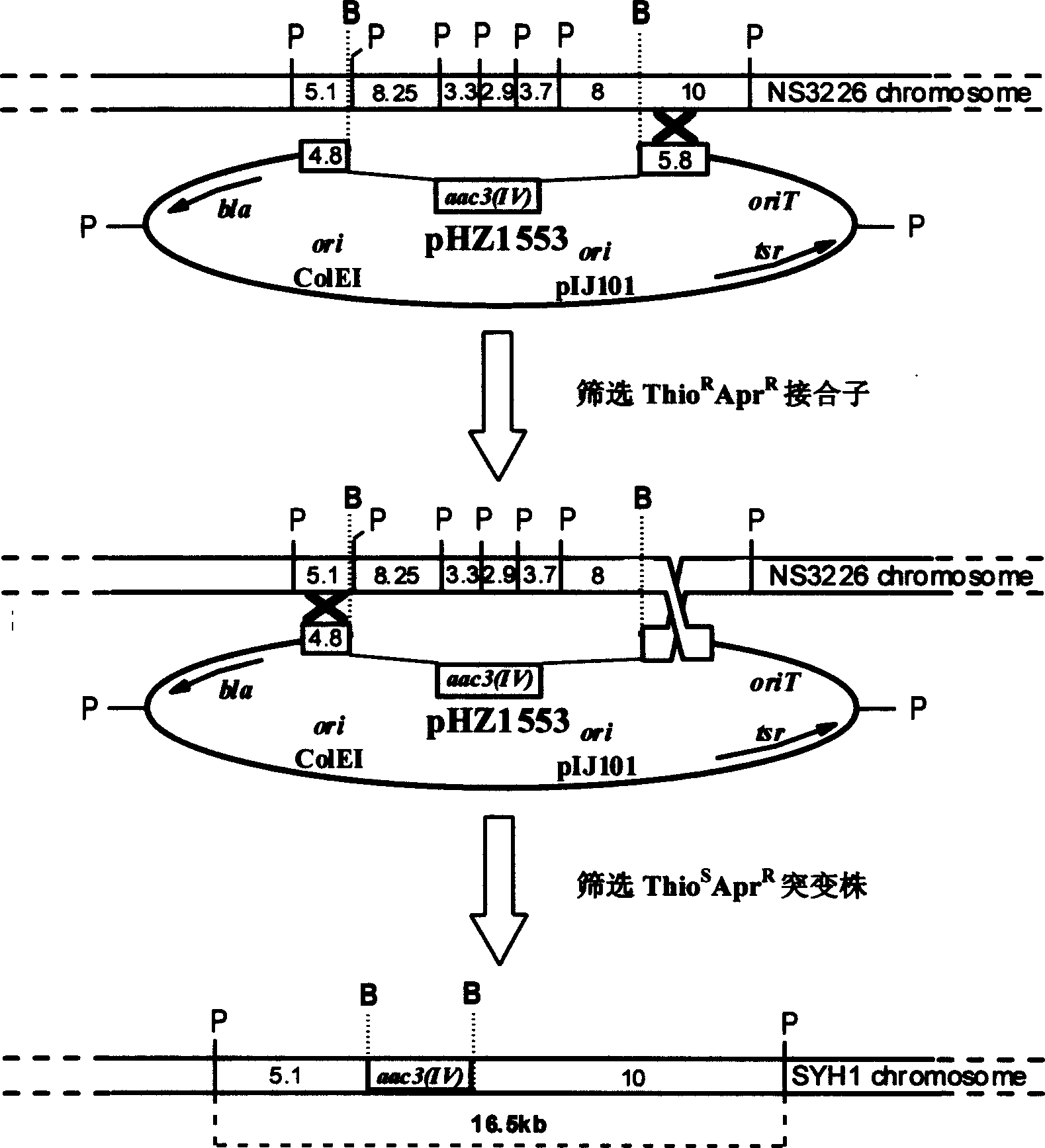
[0034] Example 2: Intergeneric conjugative transfer of pHZ1553 from E. coli to Streptomyces
[0035] In 2ml LB liquid culture medium containing ampicillin (100μg / ml), apramycin (30μg / ml), kanamycin (10μg / ml), chloramphenicol (25μg / ml) , yeast extract 5g, NaCl 5g, distilled water 1000ml, pH7.0) inoculate Escherichia coli ET12567 (pUZ8002+pHZ1553), 37 ℃ rotary culture (220rpm) 12hr, inoculate to 5ml containing the same concentration of antibiotics according to the ratio of 1:10 In LB liquid medium, rotate culture (220rpm) at 37° C. for 2.5 hr, and then wash twice with LB liquid medium. Suspend the spores of Streptomyces nanchangus as the recipient in 5ml 0.05mol / L TES buffer solution of pH 8.0, heat shock in a water bath at 50°C for 10min, cool with tap water and add an equal volume of spore pre-germination medium (Difco yeast extract 1g , Difco Casamino acids 1g, CaCl 2 0.01M (need to prepare 5M stock solution, add to yeast extract / casein amino acid solution after separate ste...
Embodiment 3
[0036] Example 3: Screening of gene knockout strains
[0037] The acquired apramycin and thiostrepton resistance (Thio R April R ) conjugative transferons, and then passed through non-resistant GS plate (soluble starch 20g, KNO 3 1g, K 2 HPO 4 0.5g, MgSO 4 ·7H 2 O 0.5g, NaCl 0.5g, FeSO 4 0.01g, 20g of agar, 1000ml of distilled water, pH7.5) After 7 days of relaxation culture, copy to GS plates containing thiostrepton (5μg / ml) and apramycin (10μg / ml) respectively, and screen the phenotype resistant to apramycin and sensitive to thiostrepton (Thiostrepton S April R ) strains, these strains may be gene knockout mutants.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com