Method and kit for detecting Shigella and ipaH pathogenicity island thereof
A Shigella and kit technology, which is applied in the field of real-time quantitative fluorescent polymerase chain reaction technology to detect Shigella contamination, can solve problems such as high cost, difficult clinical detection, PCR product contamination, etc., and achieve the effect of rapid diagnosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: the development of Shigella detection reagent
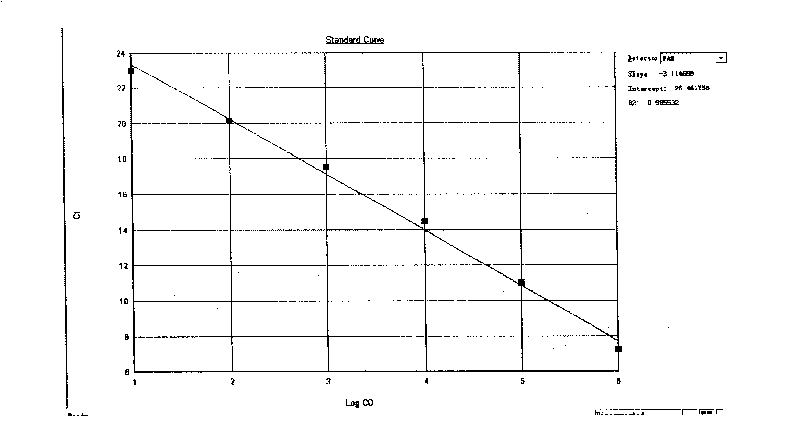
[0032] 1. Design of primers and probes: Through sequence comparison and analysis of all Shigella nucleic acid sequences already available in the Genbank database and nucleic acid sequences reported in published literature at home and abroad, the ipaH gene was selected to have no secondary structure and was highly conserved. According to the basic principles of primer-probe design, multiple pairs of primers and probes are designed by software and manually.
[0033] 2. Establishment and optimization of the reaction system
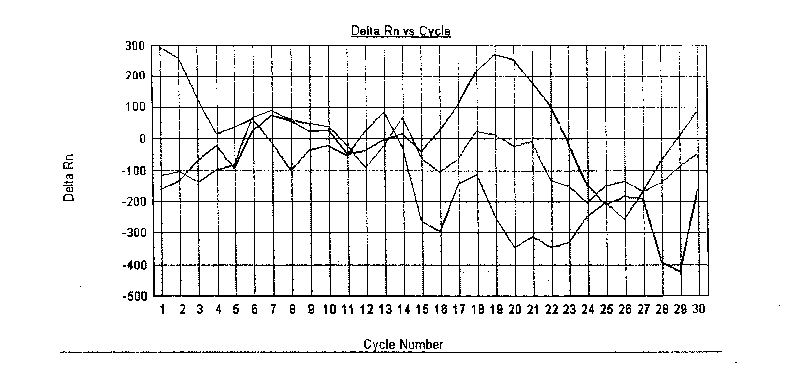
[0034] Sample preparation: the standard strain 51313 purchased by the Institute of Biological Products of the Chinese Academy of Sciences was used as a positive standard for Shigella detection; as a negative reference. The genomic DNA of the above-mentioned positive standard product and negative reference product were extracted by boiling the DNA extraction solution for use.
[0035] Scre...
Embodiment 2
[0043] Embodiment 2: Shigella detection kit and its use
[0044] 1. Prepare a kit including the following components: 2 tubes of DNA extraction solution (500 μl / tube), 20 tubes of PCR reaction solution, 1 tube of negative reference product (100 μl / tube), 1 tube of critical positive reference product (50 μl / tube) , Positive quantitative standard (50μl / tube) 4 tubes.
[0045] 2. Specimen collection, transportation and storage
[0046] 2.1 Feces: Collect mucus, pus and blood feces in the early stage of the disease (before treatment) for bedside inoculation. If the inoculation cannot be inoculated in time, it can be placed in glycerin preservation solution or card-cloth transport medium for inspection.
[0047] 2.2 Specimen storage and delivery: Specimens can be used for testing immediately, or can be stored at -20°C for testing, and the storage period is 6 months. Specimens should be transported in a 0°C ice bottle.
[0048] 3. Detection steps
[0049] Add 4 times the volume ...
Embodiment 3
[0052] Embodiment 3: Detecting the specimen for determining the type of Shigella
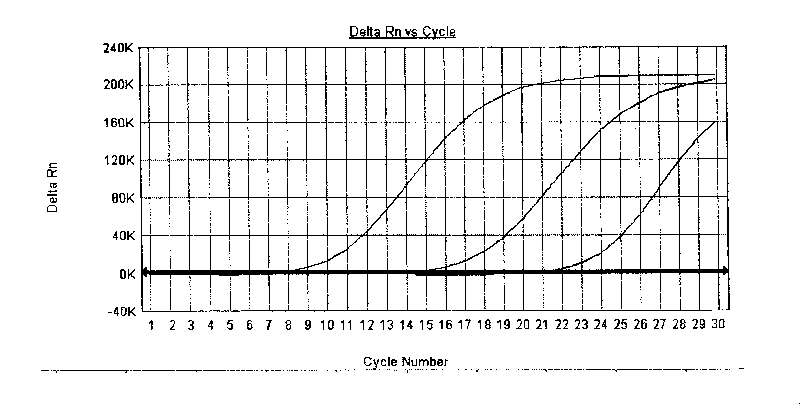
[0053] The specific steps are the same as in Example 2, except that the samples used are Shigella dysenteriae, Shigella flexneri, Shigella baumannii, and Sonnei from China Microorganism Culture Collection Center and China Institute for the Control of Pharmaceutical and Biological Products. Shigella. After nucleic acid is extracted from the specimen, the kit of the present invention is used for PCR amplification, and the instrument automatically monitors and collects changes in fluorescent signals during the amplification. Analysis after reaction finishes, application kit of the present invention all can detect the specimen of known type (referring to Figure 4 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com