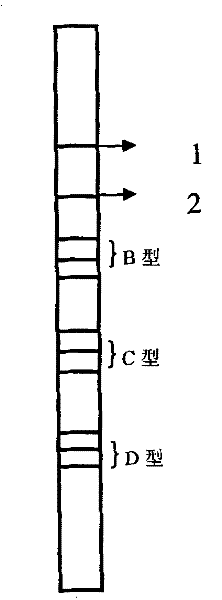
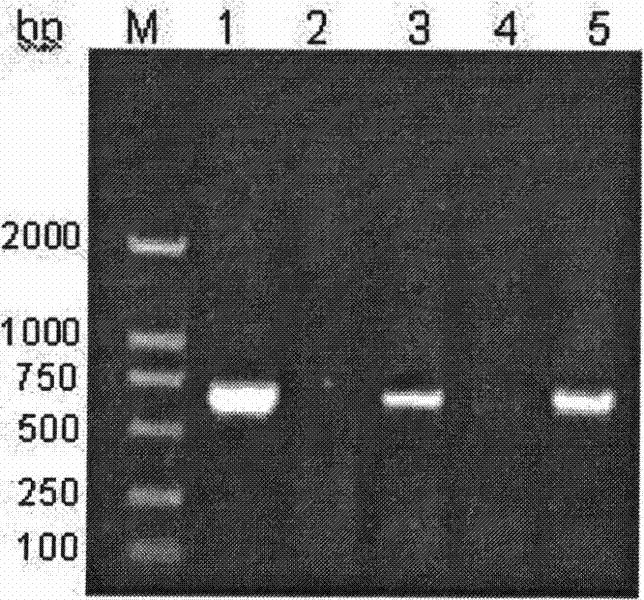
A method and kit for detecting hepatitis B virus genotype
A hepatitis B virus and genotyping technology, applied in the directions of microorganism-based methods, biochemical equipment and methods, and microorganism determination/examination, etc., can solve the problems of affecting the typing results, unable to identify specimens, complex band types, etc. To achieve the effect of enhancing detection sensitivity, improving binding efficiency, and improving signal strength
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Extracting HBV Genomic DNA from Serum
[0047] Take 200ul virus serum + 200ul 2x lysis buffer (0.02mol / L TrisHCl, 0.01mol / L EDTA, 1% SDS) + 10ul proteinase K (20mg / ml), digest at 40°C for 1.5 hours. Use phenol and phenol : Chloroform (1:1), each extracted once with chloroform, after mixing, 12000gx3min, recover the water phase, ie the upper layer. Add 1 / 10 volume of 3mol / L sodium acetate and 2 volumes of pre-cooled absolute ethanol to precipitate DNA, 12000g x 10 minutes, recover DNA, wash DNA once with 75% ethanol, discard supernatant, after drying, add 20μl pure Dissolve DNA in water, take 5 μl as template for PCR reaction, and store the rest at -20°C.
Embodiment 2
[0048] Embodiment 2PCR reaction amplifies target DNA fragment
[0049] Externally amplify 50 μl PCR system as follows:
[0050] 10×Taq amplification buffer (TaKaRa) 5 μl
[0051] MgCl 2 (25mM) 4μl
[0052] dNTP Mixture (2.5mM each) 4μl
[0053] Upstream primer (20um) 1μl
[0054] Downstream primer 1 (20um) 1μl
[0055] Taq DNA polymerase 5U / μl (TaKaRa) 0.25μl
[0056] Autoclaved distilled water 29.75μl
[0057] Template 5 μl purified HBV DNA, or 5 μl distilled water (negative control)
[0058] Put each reaction tube into the calibrated PCR instrument. Start to perform the following external amplification program: 94°C pre-denaturation for 4 minutes, then 40 cycles of 94°C for 30 seconds, 56°C for 40 seconds, 72°C for 50 seconds, and finally 72°C for 10 minutes.
[0059] After completing the above procedures, the samples should be used for electrophoresis immediately, and if there is no positive band, perform internal amplification, and store the rest at -20°C or perfo...
Embodiment 3
[0073] The preparation of embodiment 3 hybridized carrier nylon film strips
[0074] Dilute each artificially synthesized probe to a final concentration of 10 pmol / μl, and fix it on the appropriate position of the membrane strip. The preparation of the membrane strip is as follows:
[0075] (1) Add the DIG-labeled PCR product and 10 probes into the No. 1 pump and No. 2 pump of the automatic film spraying machine respectively. After spraying, clean the two pipes with distilled water and replace them again. The other two probes are sprayed with film.
[0076] (2) Set working parameters: spray volume 2 μl / cm, rail speed 6.0 cm / s, nozzle interval 5 mm.
[0077] (3) Fixing the probe: After spraying, let the film dry naturally at room temperature, perform ultraviolet cross-linking, and then bake at 120°C for 30 minutes.
[0078] (4) Cutting: After fixing, cut the film into 3mm×7cm film strips with a strip cutter along the marking line.
[0079] (5) Pre-hybridization: Add 2ml of p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com