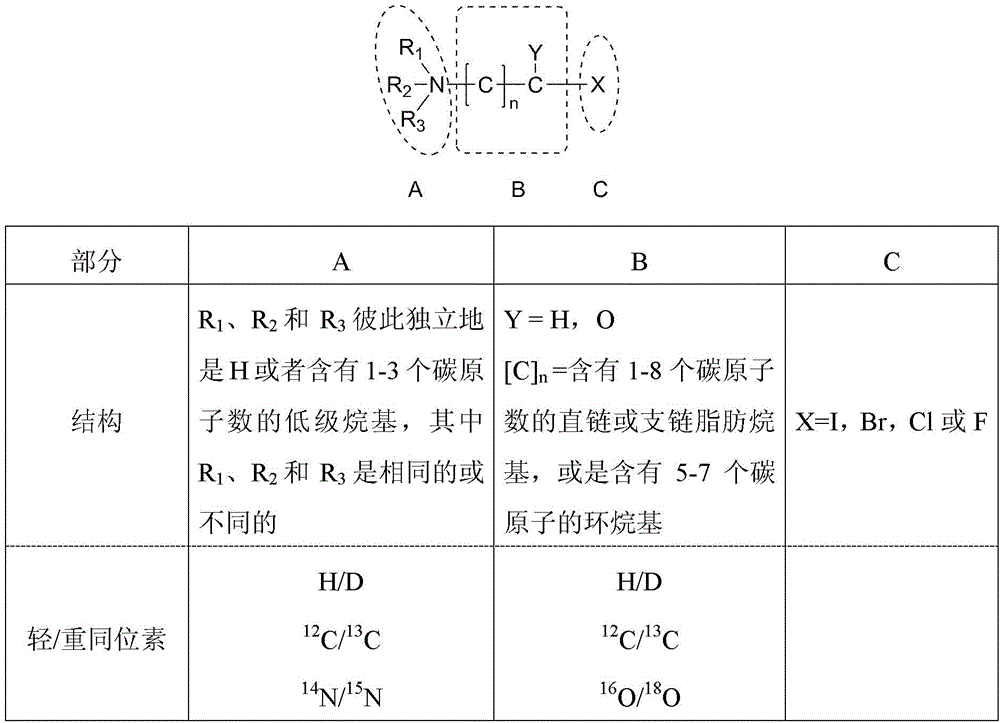
Relative quantification method based on novel mass discrepancy labels for carboxylic acid type signal molecules in biological system
A mass difference, signal molecule technology, applied in the preparation of amino compounds, chemical instruments and methods, material separation, etc., can solve problems such as difficulty in obtaining, low sample throughput, and inability to detect signal molecules, saving manpower and improving samples. The effect of flux
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
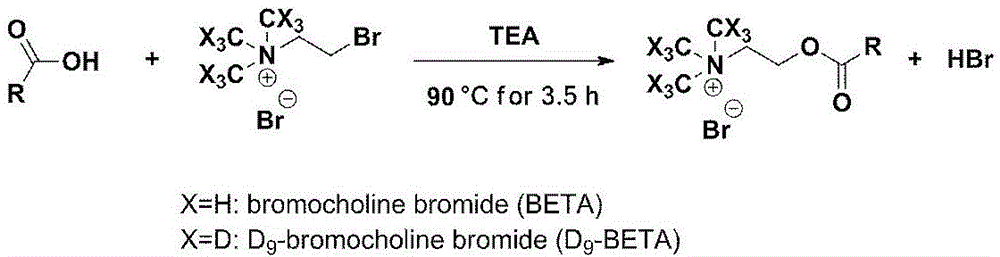
[0109] Synthesis of (2-bromoethyl)trimethylammonium bromide (BETA) 9-deuterated isotope labelled molecule D 9 -BETA
[0110] 1.12g D 9 -Trimethylamine reacts with 7.1mL 1,2-dibromoethane in a dry ice-acetone bath under the action of 6mL dry methanol;
[0111] After the reaction mixture returned to room temperature, it was stirred at room temperature for 2 days;
[0112] The formed white solid was filtered and washed with cold ether to obtain 2.64g of product D. 9 -BETA, 62% yield.
[0113] use 1 H-NMR ( 1 H-NMR), high resolution mass spectrometry (HR-MS) and high resolution tandem mass spectrometry (HR-MS / MS) analysis confirmed the synthesis of D 9 -BETA. Take TMS as internal standard, D 2 O is the solvent, 1 H-NMR (300Hz, Bruker ARX300NMR spectrometer) analysis results are as follows: 3.737(s, 4H), Figure 4 . The mass spectrometer is equipped with an electrospray ion source (ESI) (Waters Synapt TM G2 high resolution mass spectrometry), monitored in positive ion mode, the analysis ...
Embodiment 2
[0115] Relative quantitative analysis of the above 8 carboxylic acid signal molecules in 0.8 mg dry weight (DW) model plant Arabidopsis (Col-0) treated with methyl jasmonate (MeJA) and untreated for 12 days
[0116] Treatment of samples in the treatment group: Add 4 mL of 100 mM MeJA (solvent of 0.1% ethanol) to a culture dish of Arabidopsis whole seedlings grown under normal conditions for 10 days, and then put them in the greenhouse for 2 days after 10 minutes. Grind down and freeze-dry
[0117] Control sample treatment: Add 4 mL of 0.1% ethanol (blank solvent) to the culture dish of Arabidopsis thaliana whole seedlings grown for 10 days under normal conditions, and then put it in the greenhouse for 2 days after 10 minutes, and then under liquid nitrogen after sampling Grind and freeze-dry
[0118] Weigh 0.8mg dry weight (DW) samples of the control group and the treatment group, extract them with methanol and blow dry at room temperature, re-dissolve them in 150μL of acetonitrile,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com